Highly sensitive and effective tool measures how cells grow and divide
In a paper published in the journal Genes and Development on July 10, an IBS research team presents mTAIL-seq, a hyper-sensitive, upgraded tool to measure poly(A) tail length of mRNAs at the genomic scale. By revealing the dynamic poly(A) regulation during the translation of RNA into protein, the study advances our understanding of the fabric of life, from the color of your skin to how tall you will grow.
The essential three & the building blocks of life
All life on Earth requires three separate but equally essential biological molecules that serve a critical function in a cell: Proteins, DNA and RNA. Proteins are the workers; they perform diverse catalytic and structural roles within cells. Both DNA and RNA carry genetic information that's inherited from generation to generation. They are the reason why some people have black hair, others blond with blue eyes and others still with green eyes. Central dogma is a biological concept that explains how DNA and RNA interact to produce proteins. Developed in the late 50's, by British molecular biologist Francis Crick, the concept fundamentally outlines the three stages: DNA replicates its information using many enzymes after which DNA's encoded information is transcribed into RNA whereupon a variation of RNA is translated into proteins. During the transcribing process various forms of RNA are synthesized, each has an essential function to help create a template for future construction of protein. Messenger RNA (mRNA) is the architect and instructs transfer RNA (tRNA) and ribosomal RNA (rRNA) on how to correctly assemble amino acids that, when combined into chains, create proteins: the building blocks of all life.
mTAIL-seq: a highly sensitive and cost effective tool to measure poly(A) tail in eggs-to-embryos development
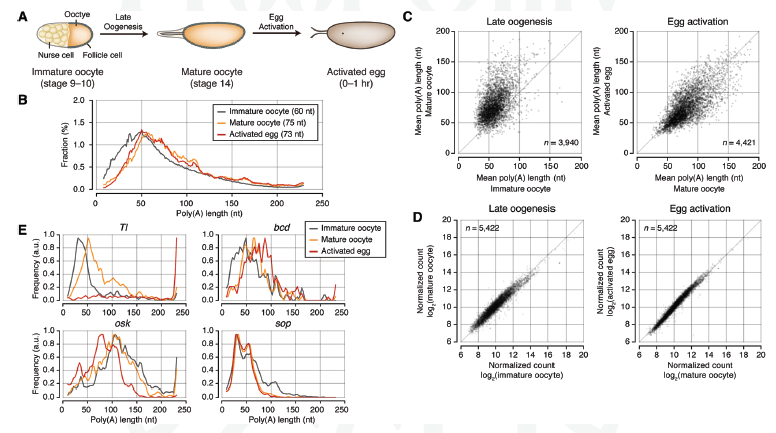
The IBS team focused on oocytes (eggs) in which maternal mRNAs are deposited in a dormant state, yet to be translated into proteins. In early embryos, where transcription is silent, the activation of the dormant mRNAs is vital for the proper production of proteins; thus a deeper understanding of the regulation of maternal mRNAs can reveal how oocytes establish the maternal characteristics of embryos. The key mechanism to controlling maternal mRNAs is the regulation of poly(A) tail: A chain booster that's added to mRNA to increase its stability and promote protein synthesis. Inside an oocyte, long poly(A) tails are added to mRNAs, allowing for them to translate and build proteins; proteins that are required for early embryonic development. The team's upgraded tool, mTAIL-seq, allows enhanced sequencing depth for mRNA at a rate of 1,000 finer than its predecessor. First author, Dr. LIM Jaechul explains further, "We used mTAIL-seq to measure poly(A) length of maternal mRNAs in oocyte-to-embryo development. From the genomic scale analysis, we found global dynamic poly(A) tail regulation without the change of mRNA abundance."
Comparing two indices for one complete picture
Ribosome profiling (RPF) data measures the efficiency of translation, from mRNA to protein, the second step in central dogma. When the RNA team compared the RPF data with data acquired from mTAIL-seq they found a strong coupling between poly(A) tail length and the translational efficiency at early embryo stage. This data, according to the team's manuscript, "suggests that regulation of poly(A) tails in oocytes shapes the maternal characteristics {translatomic landscape} of embryos, thereby directing the onset of animal development."
"The global profiling of poly(A) tails by mTAIL-seq provides a comprehensive resource for the regulation of poly(A) tails in Drosophila {fruit flies} oocyte-to-embryo development and it help us to understand how poly(A) tail of maternal mRNA affect the production of proteins at the beginning of embryonic development," said the first author Dr. Mihye LEE.
Due to the high sensitivity and low cost of mTAIL-seq not to mention its technical robustness and broad accessibility, the team think their invention will be a potent tool to improve understanding of mRNA tailing in diverse biological systems.
More information: Jaechul Lim et al. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development, Genes & Development (2016). DOI: 10.1101/gad.284802.116
Journal information: Genes & Development
Provided by Institute for Basic Science