Laser tool effective at identifying mutant listeria bacteria
A Purdue University-developed laser tool already effective in quickly detecting harmful bacteria has been shown to detect mutant varieties of listeria - and in the same amount of time.
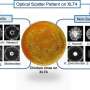
The BARDOT (pronounced bar-DOH') laser scans bacteria colonies looking for unique patterns that each bacterium makes. When the light penetrates a bacteria colony, it produces a scatter pattern that can be matched against a library of known bacteria patterns to identify a match. The system can identify bacteria such as salmonella, listeria, bacillus, vibrio and E. coli within 24 hours.
Now, Arun Bhunia, professor of food microbiology, and Atul Singh, research scientist, have shown that BARDOT (acronym for "bacterial rapid detection using optical scatter technology") can pinpoint small genetic mutations in listeria just as quickly, significantly reducing the time it would take scientists to identify those mutations in bacterial strains used for research. Their study was published in the journal Applied and Environmental Microbiology.
"This is a versatile microbiology tool, and we wanted to see if we can use it for mutant strains," Bhunia said. "This is a really powerful tool to help researchers find those mutant strains much easier on a petri plate. You can avoid the laborious techniques required to screen or detect these mutant strains."
Scientists use mutant bacteria to understand biology of pathogens and how they can combat outbreaks in food that can cause illnesses or death. Current methods of identifying mutants can take several days, whereas BARDOT can do the same work in less than a day.
Singh said he visualized the changes in bacteria using the laser system.
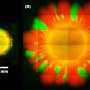
"It's like if you compare a wild type that is a normal bacteria, you get a scatter pattern. And then if you delete a certain gene, you get a new picture," he said.
The reverse was also true. By restoring the deleted gene, the BARDOT system recognized the bacteria as a regular wild type of strain.
Bhunia said his lab will continue to study BARDOT's ability to identify mutants of other bacteria and build libraries so the tool can be used for that work. He will also test the system's ability to identify mutant bacteria from natural settings such as from contaminated food.
More information: Atul K. Singh et al. Virulence Gene-Associated Mutant Bacterial Colonies Generate Differentiating 2-D Laser Scatter Fingerprints, Applied and Environmental Microbiology (2016). DOI: 10.1128/AEM.04129-15
Journal information: Applied and Environmental Microbiology
Provided by Purdue University