New insights into protein structure could change the future of biomedicine
Researchers at the University of Waterloo have discovered a new way to create designer proteins that have the potential to transform biotechnology and personalized medicines.
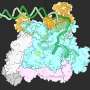
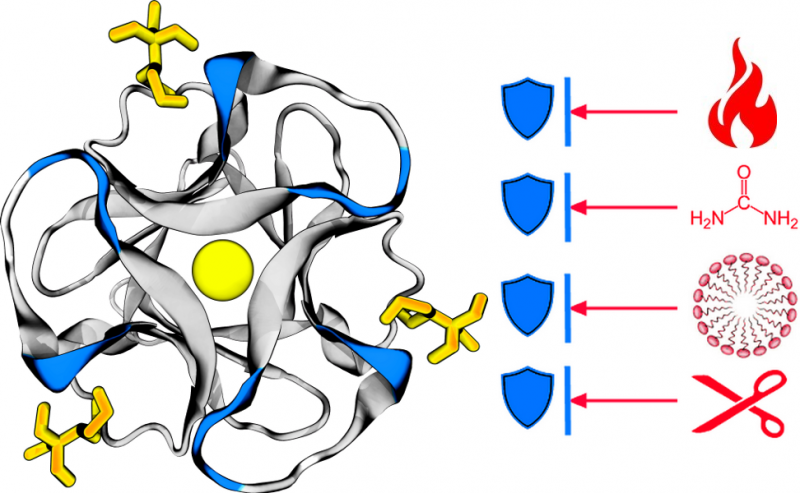
In a range of experiments Professor Elizabeth Meiering, in collaboration with colleagues from India and the United States, created a protein that can withstand a range of physiological and environmental conditions - a problem that has challenged chemists looking to create super stable, highly functional proteins.
Their results are published this month in the prestigious, peer-reviewed journal Proceedings of the National Academy of Sciences.
Protein drugs can be engineered to act like antibodies and search out specific cells. Such personalized medicines can attach only where they are needed, greatly reducing side effects in cancer and arthritis treatments. For example, the blockbuster drug Herceptin, currently used to treat several forms of breast cancer, targets HER2 growth receptor sites on the surface of cancer cells and tags the cells so the body's immune system can destroy them.
Nevertheless, designing a protein that can withstand a range of conditions is challenging and can be risky. Proteins rely on their unique structure to perform their function and one small change to that structure can lead to an allergic reaction, or even a deadly cascade immunological response.
"It's not enough to design a protein partially right—it needs to be exactly right in order for the protein to be stable and functional and for a drug to work," said Professor Meiering. "Most natural proteins aren't very stable, and chemists are finding it hard to design stability. Our work represents a real shift in how researchers can engineer and understand proteins."
Traditionally protein designers focus on either structure or function because working with each property is challenging in its own right.
Professor Meiering and her colleagues were able to incorporate both structure and function into the design process by using bioinformatics to leverage information from nature. They then analyzed what they made and measured how long it took for the folded, functional protein to unfold and breakdown.
"A protein can be energetically unstable, yet still unfold extremely slowly—meaning it remains folded and functional for a long time," said Professor Meiering, a member of the Centre for Bioengineering and Biotechnology.
Using a combination of biophysical and computational analyses, the team discovered this kinetic stability can be successfully modeled based on the extent to which the protein chain loops back on itself in the folded structure. Because their approach to stability is also quantitative, the protein's stability can be adjusted to naturally break down when it is no longer needed.
This different way of thinking will allow researchers to move forward with designing proteins with precisely controlled stability for challenging applications such as biosensors and personalized therapeutics.
More information: A. Broom et al. Designed protein reveals structural determinants of extreme kinetic stability, Proceedings of the National Academy of Sciences (2015). DOI: 10.1073/pnas.1510748112
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Waterloo