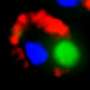
How human cells can dissolve damaging protein aggregates
Cellular repair systems can dissolve aggregated proteins and now Heidelberg researchers have successfully decoded the fundamental mechanism that is key to dissolving these protein aggregates in human cells. Their in-vitro experiments uncovered a multi-stage biochemical process in which protein molecules are dissolved from the aggregates. Researchers at the Center for Molecular Biology of Heidelberg University, the German Cancer Research Center and the Heidelberg Institute for Theoretical Studies collaborated on the project, along with other scientists from Germany, the USA and Switzerland. The results of their research were published in Nature.
Proteins in all cells – from bacteria to human – are folded in their native state. Proteins are first manufactured as long, sequential chains of amino acids and must assume a specific three-dimensional structure, i.e., fold, to be functional. This correctly folded state, or protein homeostasis, is at constant risk from external and internal influences. Damaged proteins lose their structure, unfold and then tend to clump together. "If such aggregates form, they can damage the cells and even cause the cells to die, which we see in neurodegenerative diseases such as Alzheimer's and Parkinson's, and even in ageing processes," explains Prof. Dr. Bernd Bukau, Director of the Center for Molecular Biology of Heidelberg University (ZMBH), who is also a researcher at the German Cancer Research Center (DKFZ).
Prof. Bukau explains that damaged proteins do not only clump during the ageing process. Protein aggregates can also occur through changes in the protein structure due to mutation or chemical or environmental stresses. A change in growth conditions, such as an increase in ambient temperature, can cause proteins to lose their structure and unfold. "The formation of protein aggregates in different organs of the human body is associated with a large number of diseases, including metabolic disorders," explains the ZMBH Director.
The researchers report that very little was known about how our natural defences reverse the process of protein aggregation so effectively in young healthy cells. "Dissolving protein aggregates is a critical step in recycling defective proteins and providing protection against stress-induced cell damage. We had several clues as to the main players in this process, but we didn't know exactly how it worked," says lead investigator Dr. Nadinath Nillegoda, a member of Prof. Bukau's team. The researchers succeeded in identifying a previously unknown, multi-component protein complex that efficiently solubilizes stress-induced protein aggregates in vitro.
This complex consists of molecular folding helpers, the chaperones, which in this case belong to the heat shock protein 70 (Hsp70) class. These are proteins that aid other proteins in the folding process. The Heidelberg researchers also studied the co-chaperones that regulate Hsp70 activity in the protein complex. According to Prof. Bukau, the co-chaperones of the so-called J-protein family are key, in that they "lure" the Hsp70 folding helpers to the protein aggregates and activate them precisely at their target. "The key finding of our work is that two types of these J-proteins must dynamically interact to maximally activate the Hsp70 helper proteins to dissolve the protein aggregates. Only this launches the potent cellular activity to reverse these aggregates."
Scientists from the Heidelberg Institute for Theoretical Studies (HITS) performed the computational data analysis for this research. For the experimental design and integrating the data from a range of experiments, they developed a special modelling methodology for protein-protein docking to simulate the formation of chaperone complexes. HITS research group leader Prof. Dr. Rebecca Wade, who also conducts research at the ZMBH, notes that this molecular-level modelling was essential for understanding the dynamic interactions underlying the coordinated activity of the two types of J-proteins in the chaperone complex.
According to Prof. Bukau, now research is faced with the challenge of understanding the physiological role and the potential of the newly discovered mechanism well enough to apply these findings from basic research and develop novel strategies for therapeutic interventions.
More information: "Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation." Nature (published online 5 August 2015), DOI: 10.1038/nature14884
Journal information: Nature
Provided by Heidelberg University