September 9, 2015 report
Label-free technique that images DNA in vivo
(Phys.org)—A group of researchers from Harvard University report being able to observe DNA dynamics during cell division in vivo using time-lapse stimulated Raman scattering microscopy and without using fluorescent labels. Their work appears in the Proceedings of the National Academy of Sciences.
To really understand what is happening during cellular processes, including cellular malfunctioning as in cancer, there is a need to peer inside the cell without disrupting any of the cellular processes. Typically if scientists want to look at large spools of DNA, known as chromosomes, they would need to fluorescently label the DNA. This approach is invasive and may alter the native environment of the cell.
Furthermore, in medicine, histological diagnoses are typically done by examining stained tissue biopsies. In this study, Fa-Ke Lu, Srinjan Basu, Vivien Igras, Mai P. Hoang, Minbiao Ji, Dan Fu, Gary R. Holtom, Victor A. Neel, Christian W. Freudiger, David E. Fisher, and X. Sunney Xie present a label-free technique that forgoes staining and could potentially offer a non-invasive way to diagnose skin cancer.
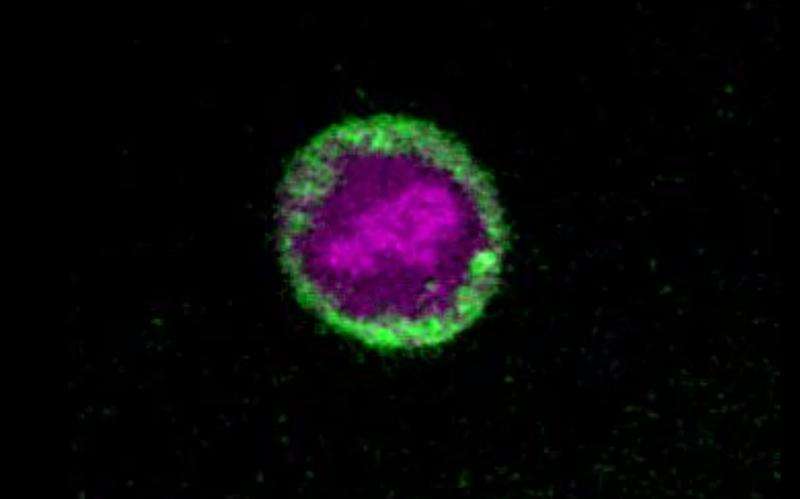
Stimulated Raman scattering (SRS) microscopy is a type of imaging technique that looks at the vibrational frequencies of chemical bonds. Different types of bonds will have different frequencies based on the surrounding molecular environment. For example, a C-H bond on a DNA molecule (2,956 cm-1) is going to have a slightly different vibrational frequency compared to a C-H bond on proteins (2,931 cm-1) or lipids (2,854 cm-1). Unlike traditional Raman spectroscopy, SRS is able to obtain data on a sample rapidly, allowing for real-time, in vivo studies. By looking at these C-H stretching vibration regions and conducting a linear decomposition of the images, Lu, et al. were able to map the content and distribution of DNA, proteins, and lipids within the cell, allowing them to observe the cell division process in a label-free manner.
To prove that their technique would work, they analyzed samples of synthetic DNA, BSA (protein) and oleic acid (lipid), individually and collectively in a cellular pellet. They measured the three Raman vibrational shifts and calculated the distribution of DNA, protein and lipid using linear decomposition. This study confirmed not only that their technique worked, but that it is also highly accurate.
The next step was to test their technique in biological samples. Lu, et al. used their SRS technique to look at cell division in HeLa cells. They first looked at cells in the first stage of mitosis, the prophase, and were able to reconstruct a 3D distribution of DNA, lipids, and protein, showing a high concentration of DNA in the nucleus, lipids predominantly in the cytoplasm, and proteins scattered throughout the entire cell. They then imaged cells in at the interphase stage of mitosis and were able to discern the chromatin structures in the nuclei. Time-lapse SRS allowed Lu, et al. to observe the transition from metaphase to anaphase.
In vivo studies were done on the skin of mice treated with TPA, a chemical that promotes cell division. Lu, et al. were able to discern each stage of the cell cycle as described before. Additionally, they were able to look at chromosomal migration in cancer cells from immune-deficient mice treated with human cancer cells. These results then lead Lu, et al. to use time-lapse SRS to study cell cycle kinetics. By understanding the rate at which cells divide, researchers can discern important factors, such as the aggressiveness of a cancer. They found that in TPA-treated mice, cell mitotic activity peaked at 18 hours and by 24 hours had decreased. This was the first time that a mitotic rate was reported in vivo in a quantitative manner.
The last step was to test their label-free SRS technique with human cancer cells to see if it is a viable technique for histologic diagnosis. To ensure the accuracy of their technique, they first imaged a tissue sample using traditional staining and compared it to their SRS technique. They then imaged fresh human skin cancer tissue from three surgical cases of squamous cell carcinoma, the second most common form of skin cancer. They found an increased number of mitotic features, which translates into increased cell division and proliferation, hallmarks of cancer cells.
Their results demonstrate that their label-free SRS imaging method is comparable to traditional staining methods for histological diagnosis. Additionally, because SRS allows for the observation of nucleic acids, specifically, researchers can do quantitative studies of mitotic kinetics within a tumor cell.
According to co-author Professor Xie, "SRS imaging could be particularly relevant for in vivo counting of the mitotic rate used in human skin cancer diagnosis. We expect that SRS may not only speed up surgical procedures by on-site label-free imaging of tumor tissue with margins, but it could also have the potential for in vivo noninvasive detection and progress evaluation of skin lesions in real time."
This technique holds much promise for being used both as a non-invasive method for skin cancer diagnosis, and as a quick evaluation of the aggressiveness of cancer cells after excision.
Co-author Dr. David Fisher adds that "this remarkable methodology provides high resolution images of cells and their nuclei within their natural biological context, providing a novel means of tracking their behavior over time. SRS is a particularly valuable tool for the evaluation of cancer cells."
More information: "Label-free DNA imaging in vivo with stimulated Raman scattering microscopy" PNAS, Fa-Ke Lu, DOI: 10.1073/pnas.1515121112
Abstract
Label-free DNA imaging is highly desirable in biology and medicine to perform live imaging without affecting cell function and to obtain instant histological tissue examination during surgical procedures. Here we show a label-free DNA imaging method with stimulated Raman scattering (SRS) microscopy for visualization of the cell nuclei in live animals and intact fresh human tissues with subcellular resolution. Relying on the distinct Raman spectral features of the carbon-hydrogen bonds in DNA, the distribution of DNA is retrieved from the strong background of proteins and lipids by linear decomposition of SRS images at three optimally selected Raman shifts. Based on changes on DNA condensation in the nucleus, we were able to capture chromosome dynamics during cell division both in vitro and in vivo. We tracked mouse skin cell proliferation, induced by drug treatment, through in vivo counting of the mitotic rate. Furthermore, we demonstrated a label-free histology method for human skin cancer diagnosis that provides comparable results to other conventional tissue staining methods such as H&E. Our approach exhibits higher sensitivity than SRS imaging of DNA in the fingerprint spectral region. Compared with spontaneous Raman imaging of DNA, our approach is three orders of magnitude faster, allowing both chromatin dynamic studies and label-free optical histology in real time.
Journal information: Proceedings of the National Academy of Sciences
© 2015 Phys.org