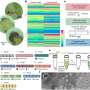
A biological escape room: Physicists get outside glimpse of protein aggregates that cause diabetes
Researchers from FOM Institute AMOLF have, for the first time ever, managed to make an image of the exterior structure of a protein aggregate that causes diseases like Alzheimer and type-2 diabetes. They discovered that the exterior surface of these aggregates are structured in very messy way – much unlike their neatly arranged interiors. The researchers hope their work will provide more insight in the way the aggregates damage nearby cells. The research findings were published online in the journal Small on 7 May, 2015.
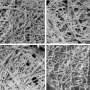
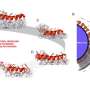
The protein aggregates studied by the researchers consist of long, folded proteins. Researchers have been familiar with the interior structure of these aggregates (formally known as amyloid fibrils) for quite a long time. However, with conventional microscopes it always remained impossible to see the protein structures at the exterior surface of the aggregates. In retrospect, it was as if the researchers were locked inside a house. They could thoroughly study the interior of the house, but as long as they didn't have the key, they were unable to step out and look at its façade on the outside. The breakthrough came when the researchers devised that 'key' by combining two different techniques: microscopy and spectroscopy. This allowed them to finally study the exterior 'brickwork'.
Messy
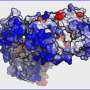
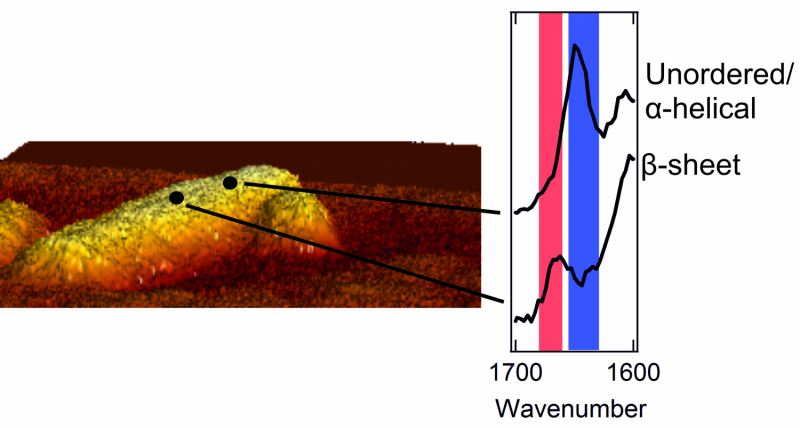
Once they could catch a glimpse of the exterior of the protein aggregates, the researchers were in for a surprise. The interior of the aggregates is strictly organized. The proteins are neatly arranged in a so called beta sheet structure, which forms a twisted, pleated sheet. However, they found that the exterior of the aggregate has a much messier and chaotic structure. That is, the exterior proteins form a disorganized, irregular pattern. Here and there, there are patches that form a beta sheet structure, but in other places this regularity is nowhere to be found. The researchers even found structural differences from one nanometre to the next, on one and the same fibril.
The researchers also noticed that each fibril's exterior structure is very different from the next. One fibril may contain many patches with a beta sheet structure, while another lacks these regular patches almost completely. Moreover, the amino acids that can be found on the exterior differ for every fibril as well.
The messy exterior is in direct contact with the membranes of neighbouring cells. In this way, its structure plays a crucial role in damaging these cells. This type of cell damage is commonly found in patients with type-2 diabetes or Alzheimer's disease.
The 'key' that opened the doors
The AMOLF researchers collaborated with their colleagues at the Utrecht University, the Max Planck Institute for Polymer Research in Mainz, and the Leibniz Institute for Photonic Technology in Jena to develop a new way to view the aggregates' exterior's protein structure. The key turned out to be a combination of spectroscopy and atomic force microscopy.
An atomic force microscope consists of a sharp tip (probe) that moves over an object's surface. This allows the microscope to explore an amyloid fibril on an atomic scale and make a relief map. Meanwhile, the spectrometer measures the so-called Raman spectrum for every pixel of the map. These spectra give information about the way the proteins are folded, and the presence of specific amino acids on the aggregates' surface. The signals measured by the spectrometer are usually weak, but a metallic layer on the tip of the microscope amplifies them greatly.
Images of the amyloid fibrils' exterior had been made before, but the team now imaged the way the proteins were folded as well. First author Corianne van den Akker: "By combining microscopy and spectroscopy, we managed to link each bit of structure to a location on the amyloid's exterior. In this way, we could make a picture with nanoscale resolution, as well as determine the structure at each location. That's the unique feat of the technique we used."
The next step
With the newly gained knowledge, it will be possible to measure the structure of amyloids that have formed on the exterior of living cells as well. The technique can also be used to test medication that aims to prevent or dissolve protein aggregates.
More information: Nanoscale heterogeneity of the molecular structure of individual hIAPP amyloid fibrils revealed with tip-enhanced Raman spectroscopy, Corianne C. van den Akker, Tanja Deckert-Gaudig, Michael Schleeger, Krassimir P. Velikov, Volker Deckert, Mischa Bonn, Gijsje H. Koenderink, Small, published online on 7 May 2015
Journal information: Small
Provided by Fundamental Research on Matter (FOM)