X-rays unlock a protein's SWEET side
Sugar is a vital source of energy for both plants and animals alike.
Understanding just how sugar makes its way into the cell could lead to the design of better drugs for diabetes patients and an increase in the amount of fruits and vegetables farmers are able to grow. Stanford University researchers have recently uncovered one of these "pathways" into the cell by piecing together proteins slightly wider than the diameter of a strand of spider silk.
To determine the size, shape and orientation of one of the newest (and smallest) of these proteins, the sugar transporter, researchers turned to the North Eastern Collaborative Access Team's (NE-CAT) and the General Medical Sciences and Cancer Institute's Structural Biology (GM/CA) Facility research facilities at the Advanced Photon Source at the U.S. Department of Energy's Argonne National Laboratory.
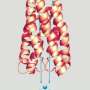
These transporters, called SWEETs, are proteins that act as pathways for sugar molecules to cross the cell membrane, where they can be used as energy to power the cell. They exist in everything from bacteria to plants to animals, though their functionality may differ for each.
The sugar you sprinkle on your breakfast cereal in the morning would never fit through one of these pathways. The sugars moving through SWEETs are the size of a molecule, invisible to the naked eye.
Like a canal's gate and lock system for boats, SWEETs allow sugar molecules to enter this pathway, closing one gate and opening another, in three different stages.
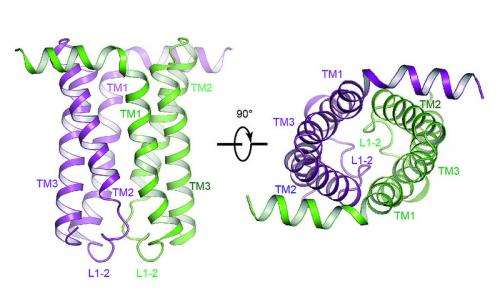
"This is the first time we were able to view the transporter's structure and begin to understand the mechanism by which it functions," said Stanford University researcher Liang Feng. "The machinery changes shape dramatically in order for molecules to move between the outside and the inside of the cell."
Previous research indicated that SWEETs transport sugar molecules, but until now, researchers did not know how the transporter did its job. To discover this, they had to piece the structure together, atom by atom.
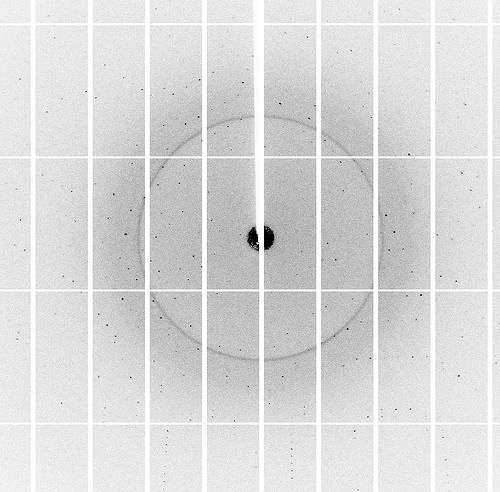
Beamlines at the NE-CAT and GM/CA, both housed at the APS, use high-powered X-rays to determine the three dimensional structure of complex biological structures, from DNA to viruses to proteins at the nanoscale. These must all be made into crystal form before going under the X-rays.
"We help scientists 'solve their structures' by providing specialized equipment and support," said NE-CAT staff scientist and macromolecular crystallographer Kay Perry. "Sometimes the crystals are really small, or there's a quality issue, or maybe crystals are growing on top of other crystals—there are a bunch of different issues for researchers, but solving them is our specialty."
The APS circles electrons near light speed around the ring using powerful magnets. Working in concert with the APS, NE-CAT and GM/CA accept high intensity X-rays generated by these magnets and focus them through a series of mirrors into a micro-beam. Specialized equipment on these beamlines allow scientists to examine very small micrometer-sized samples, like those grown by Feng's lab.
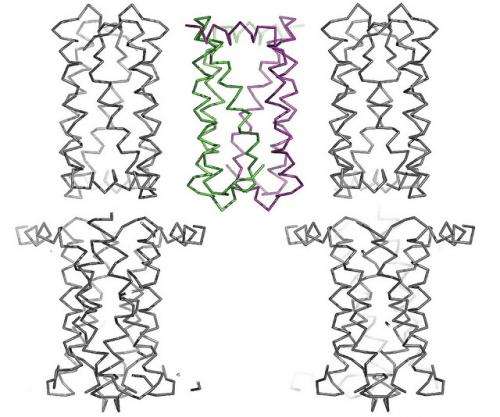
"The APS has one of the brightest beams in the world, making it able to create extremely high-resolution images," said Perry. "It worked out really well because Liang wants to see the individual bonds between atoms, and we were able to do that for him."
In collaboration with Wolf Frommer, director of Carnegie Institution for Science's Plant Biology Department, Feng's group has successfully captured SWEETs in two different shapes. Now, the two look forward to capturing even more and creating a deeper understanding of the mechanisms by which these complex machines function at the atomic level.
"Sugar is such an important source of energy for organisms, from the smallest bacteria to plants to human beings," said Feng. "This research can potentially lead us to a number of applications, from enhancing plant productivity to understanding about how animals metabolize sugar. These are just the first steps."
This research, "Structures of bacterial homologues of SWEET transporters in two distinct conformations," appeared in the September 3 issue of Nature.
More information: "Structures of bacterial homologues of SWEET transporters in two distinct conformations." Yan Xu, Yuyong Tao, Lily S. Cheung, Chao Fan, Li-Qing Chen, Sophia Xu, Kay Perry, Wolf B. Frommer & Liang Feng Nature (2014) DOI: 10.1038/nature13670
Journal information: Nature
Provided by Argonne National Laboratory