Organelles join mitochondria in signalling pathways creation
Scientists are a step closer to understanding mitochondrial pathways on a molecular level.
Research has explored factors regulating the processes and signalling pathways of metabolic and biosynthetic machinery in plant mitochondria.
Mitochondria are important hubs for signalling in plant cells; sensing other external and internal signals which integrate and optimise growth with stress responses and energy metabolism.
The study from the University of WA's Australian Research Council Centre of Excellence in Plant Energy Biology used Arabidopsis thaliana as the plant model.
Post-doctoral research associate Dr Sophia Ng, one of the study's authors, says Arabidopsis thaliana provides several advantages including a small genome size, fully sequenced genome DNA, short lifespan and lots of transfer-DNA insertion mutants that are available commercially.
"Mitochondrial retrograde signalling is more complicated than we used to think," she says.
"Pathways are originated from mitochondria directly to the nucleus.
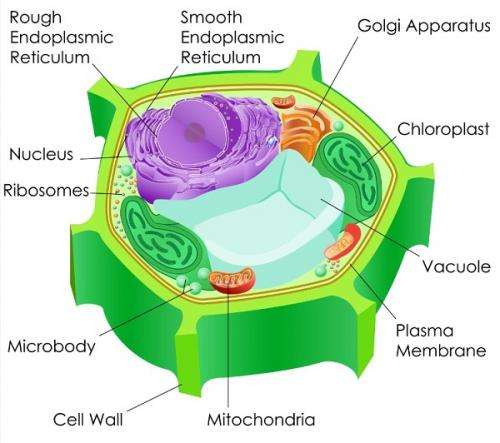
"But more studies have found that many of these signalling pathways are overlapping with pathways originated from other organelles, such as chloroplasts.
"It is proposed that signals from mitochondria and chloroplasts are able to converge into one single signal—a single regulator is activated and it then sends the retrograde signal to the nucleus."
Dr Ng says this mechanism could possibly increase the efficiency of the signalling in the cell because the convergence of a single signal works as a master switch, coordinating several organelles that are functionally correlated.
Transcription factors found in mitochondrial dysfunction
In the search for mitochondria retrograde regulators through a forward genetics screen, the research team identified several transcription factors (proteins that bind to specific DNA sequences) that are activated during mitochondrial dysfunction.
"These transcription factors, ANAC017 and ANAC013 belong to NAC family proteins that are plant-specific transcription factors," Dr Ng says.
NAC refers to the first letters of three different genes NAM (no apical meristem), ATAF (Arabidopsis transcription activation factor) and CUC (cup-shaped cotyledon).
Researchers expected to find the ANAC017 transcription factor in the nucleus but, using green fluorescent protein-targeting assays, discovered it was actually located in the endoplasmic reticulum (ER).
"When ANAC017 is activated – when the mitochondria function is perturbed – it will then move from the ER to the nucleus and bind to target genes," Dr Ng says.
"It is expected that there are many regulators in the mitochondrial retrograde pathways and what we found has added to this list."
More information: "Anterograde and Retrograde Regulation of Nuclear Genes Encoding Mitochondrial Proteins During Growth, Development and Stress." Ng S, De Clercq I, Aken OV, Law SR, Ivanova A, Willems P, Giraud E, Breusegem FV, Whelan J. Mol Plant. 2014 Apr 7. [Epub ahead of print]. www.ncbi.nlm.nih.gov/pubmed/24711293
Provided by Science Network WA