Untying DNA knots
Structural biologists at the Friedrich Miescher Institute for Biomedical Research have resolved the 3D structure of a protein machine that plays an important part in the maintenance of genomic stability. They have revealed how one unit of the machine, RMI, modulates the workings of an enzyme, topoisomerase IIIα (TopIIIα), thus allowing double Holliday junctions – key intermediates in DNA repair – to be disentangled. Their results have been published in Nature Structural & Molecular Biology.
The dissolvasome, as its name suggests, helps to remove otherwise harmful DNA intermediates by untying structural knots – so-called double Holliday junctions – that arise during DNA replication and repair. It is composed of three central proteins – a topoisomerase (TopIIIα), which relieves topological stress in DNA; BLM, a strand-separating helicase; and RMI1, a small helper protein. Mutations in any of the dissolvasome subunits result in gross chromosomal instability, but it has not been clear how the protein machine's components work together to catalyze the untangling of DNA knots.
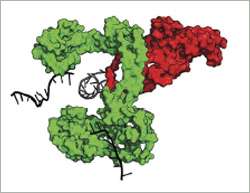
The group led by Nicolas Thomä at the Friedrich Miescher Institute for Biomedical Research has now resolved the 3D structure of RMI bound to human TopIIIα and shown how RMI modulates the activity of the dissolvasome.
"We see that RMI lines an area of TopIIIα that functions as a gate through which one strand of the DNA passes during the resolution of the double Holliday junction", explains Thomä. "RMI stabilizes the TopIIIα gate and favors an open state. This gives one of the DNA strands sufficient time to pass – a crucial step in untying the knot."
Maintaining the integrity of the DNA is vital for every cell, and defects in the protein machines that control genomic stability invariably lead to disease. In the case of the dissolvasome, mutations in the helicase BLM have been associated with Bloom syndrome – a rare genetic disease characterized by a predisposition to develop cancer.
Thomä comments: "As we start to understand in 3D how proteins in these large multi-protein complexes interact and work, we gain important insights into the regulation of these complexes. This can also be valuable for drug discovery. In our experiments, we saw for the first time how RMI1 provides new functionality to an enzyme, the topoisomerase. This illustrates why proteins operate in large assemblies – it provides them with the special bells and whistles needed to carry out their specific tasks."
More information: Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID, Thomä NH (2014) "Structural and mechanistic insight into Holliday-junction dissolution by Topoisomerase IIIα and RMI1" Nature Structural & Molecular Biology (2014) DOI: 10.1038/nsmb.2775
Journal information: Nature Structural & Molecular Biology