May 20, 2015 report
Researchers use CRISPR to create 'kill switch' for GMOs

Bob Yirka
news contributor
(Phys.org)—A pair of researchers at MIT has developed what amounts to a "kill switch" for genetically modified organisms (GMOs). In their paper published in the journal Nature Communications, Brian Caliando and Christopher Voigt describe the process they developed and how it might impact the development and use of GMOs.
GMOs have been in the news a lot of late, due to the fear by some that food produced in such a way might be harmful. Others fear that modified bacteria or viruses that make their way into the environment could prove disastrous. In this new effort, the two researchers describe a technique for adding a new functionality to GMOs—an ability to self destruct should they find themselves in the wrong environment—and central to their work is CRISPR, a means of using chemicals to modify gene fragments.
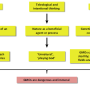
CRISPR allows for snipping gene segments and replacing them with other segments, which is the central idea behind GMOs. But CRISPR can also be used to snip out gene segments without replacing them, and that is the part of the system that Caliando and Voigt sought to exploit. Their idea is to add extra abilities to GMOs, such as the ability to recognize a certain sugar. The GMO can be further programmed to launch a secondary part of the CRISPR system when such a recognition occurs, and that secondary part would involve snipping out the segment that caused the GMO to be modified in the first place, returning it to its natural state, and killing it also if desired.
As an example, the researchers modified an already modified E. coli sample to cause it to recognize arabinose molecules—when it did so, it snipped out the parts of the DNA that had been inserted and set off a sequence of events that led to its own death. The team reports that after two hours, approximately 99 percent of those used in the test were dead.
The researchers suggest their technique could be used by companies that make GMOs in two ways. The first would be to prevent GMOs from disrupting the natural environment while the second would be to protect trade secrets—if the gene sequences that have been inserted into an organism are snipped out before it dies, others that obtain a sample would not be able to see what genetic modifications had been made.
Written for you by our author Bob Yirka—this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a donation (especially monthly). You'll get an ad-free account as a thank-you.
Publication details
Targeted DNA degradation using a CRISPR device stably carried in the host genome, Nature Communications 6, Article number: 6989 DOI: 10.1038/ncomms7989
Abstract
Once an engineered organism completes its task, it is useful to degrade the associated DNA to reduce environmental release and protect intellectual property. Here we present a genetically encoded device (DNAi) that responds to a transcriptional input and degrades user-defined DNA. This enables engineered regions to be obscured when the cell enters a new environment. DNAi is based on type-IE CRISPR biochemistry and a synthetic CRISPR array defines the DNA target(s). When the input is on, plasmid DNA is degraded 108-fold. When the genome is targeted, this causes cell death, reducing viable cells by a factor of 108. Further, the CRISPR nuclease can direct degradation to specific genomic regions (for example, engineered or inserted DNA), which could be used to complicate recovery and sequencing efforts. DNAi can be stably carried in an engineered organism, with no impact on cell growth, plasmid stability or DNAi inducibility even after passaging for >2 months.
Journal information: Nature Communications
© 2015 Phys.org