Bacterial armor holds clues for self-assembling nanostructures

Imagine thousands of copies of a single protein organizing into a coat of chainmail armor that protects the wearer from harsh and ever-changing environmental conditions. That is the case for many microorganisms. In a new study, researchers with the U.S. Department of Energy (DOE)'s Lawrence Berkeley National Laboratory (Berkeley Lab) have uncovered key details in this natural process that can be used for the self-assembly of nanomaterials into complex two- and three-dimensional structures.
Caroline Ajo-Franklin, a chemist and synthetic biologist at Berkeley Lab's Molecular Foundry, led this study in which high-throughput light scattering measurements were used to investigate the self-assembly of 2D nanosheets from a common bacterial surface layer (S-layer) protein. This protein, called "SbpA," forms the protective armor for Lysinibacillus sphaericus, a soil bacterium used as a toxin to control mosquitoes. Their investigation revealed that calcium ions play a key role in how this armor assembles. Two key roles actually.
"Calcium ions not only trigger the folding of the protein into the correct shape for nanosheet formation, but also serve to bind the nanosheets together," Ajo-Franklin says. "By establishing and using light scattering as a proxy for SbpA nanosheet formation, we were able to determine how varying the concentrations of calcium ions and SbpA affects the size and shape of the S-layer armor."
Details on this study have been published in the journal ACS Nano in a paper titled "Ion-Specific Control of the Self-Assembly Dynamics of a Nanostructured Protein Lattice." Ajo-Franklin is the corresponding author. Co-authors are Behzad Rad, Thomas Haxton, Albert Shon, Seong-Ho Shin and Stephen Whitelam.
In the microbial world of bacteria and archaea, external threats abound. Their surrounding environment can transition from extreme heat to extreme cold, or from highly acidic to highly basic. Predators are everywhere. To protect themselves, many bacteria and archaea encase themselves within a shell of S-layer proteins. While scientists have known about this protective coating for many years, how it forms has been a mystery.
Ajo-Franklin and her colleagues have been exploring self-assembling proteins as a potential means of creating nanostructures with complex structure and function.
"At the Molecular Foundry, we've gotten really good at making nanomaterials into different shapes but we are still learning how to assemble these materials into organized structures," she says. "S-layer proteins are abundant biological proteins known to self-assemble into 2D crystalline nanosheets with lattice symmetries and pore sizes that are about the same dimensions as quantum dots and nanotubes. This makes them a compelling model system for the creation of nanostructured arrays of organic and inorganic materials in a bottom-up fashion."
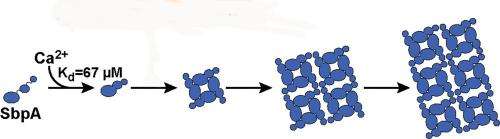
In this latest study, light-scattering measurements were used to map out diagrams that revealed the relative yield of self-assembled nanosheets over a wide range of concentrations of SbpA and calcium ions. In addition, the effects of substituting manganese or barium ions for calcium ions were examined to distinguish between a chemically specific and generic divalent cation role for the calcium ions. Behzad Rad, the lead author of the ACS Nano paper, and co-workers followed light-scattering by light in the visible spectrum. They then correlated the signal to nanosheet formation by using electron microscopy and Small Angle X-ray Scattering (SAXS), a technology that can provide information on molecular assemblies in just about any type of solution. The SAXS measurements were obtained at the "SIBYLS beamline (12.3.1) of Berkeley Lab's Advanced Light Source.
"We learned that only calcium ions trigger the SbpA self-assembly process and that the concentrations of calcium ions inside the cell are too low for nanosheets to form, which is a good thing for the bacterium," says Rad. "We also found that the time evolution of the light scattering traces is consistent with the irreversible growth of sheets from a negligibly small nucleus. As soon as five calcium ions bind to a SbpA protein, the process starts and the crystal grows really fast. The small nucleus is what makes our light-scattering technique work."
Ajo-Franklin, Rad and their co-authors believe their light-scattering technique is applicable to any type of protein that self-assembles into 2D nanosheets, and can be used to monitor growth from the nanometer to the micrometer scales.
Given the rugged nature of the S-layer proteins and their adhesive quality - bacteria use their S-layer armor to attach themselves to their surroundings - there are many intriguing applications awaiting further study.
"One project we're exploring is using SbpA proteins to make adhesive nanostructures that could be used to remove metals and other contaminants from water," Ajo-Franklin says. "Now that we have such a good handle on how SbpA proteins self-assemble, we'd like to start mixing and matching them with other molecules to create new and useful structures."
Provided by Lawrence Berkeley National Laboratory