Public computational biology web service gains popularity
"Many heads are better than one" is true for computers as well as humans. For years, scientists have connected computers into clusters and grids all over the world, crowdsourcing computational power to perform calculations that are too difficult for any single computer. Well-known projects that use public computers for data crunching include SETI@home, which analyzes radio telescope data in the search for extraterrestrial intelligence, and Folding@home, which studies protein folding to help develop drugs and combat disease.
In 2010, a team of researchers from the University of Maryland and the University of Arizona launched MolecularEvolution.org, a computing service that stands out from the crowd by giving back to said crowd. Public participants aren't only donating their computer power, but can access the network for their own projects. MolecularEvolution.org gives anyone with a computer terminal access to a worldwide grid for computational biology, known as The Lattice Project, which is built on the Berkeley Open Infrastructure for Network Computing. The grid currently offers a service called GARLI (Genetic Algorithm for Rapid Likelihood Inference), which is designed to reconstruct and predict the genetic relationships between biological samples.
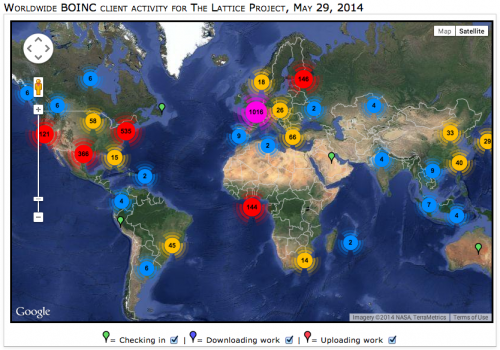
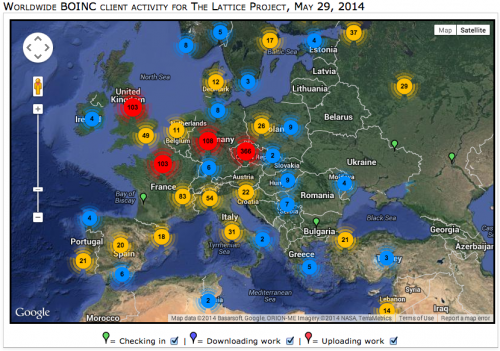
To date, over 17,000 volunteers from 146 countries have run computational biology analyses from The Lattice Project on their computers. Researchers have used the grid system to study diverse topics ranging from simulating pandemic flu risk to tracing the lineage of ancient moth species and have published 61 peer-reviewed journal articles and conference papers detailing their findings.
"Computational biology often requires more resources than any single researcher or institution could easily obtain and such resources are certainly beyond the reach of most public users who lack access to institutional equipment," explained Michael Cummings, UMD associate professor of biology and the principal investigator developing MolecularEvolution.org and The Lattice Project.
Early contributors and users of The Lattice Project include groups at UMD, Coppin State University and the Smithsonian Institution, which maintain institutional computers as part of the grid. The Lattice Project also runs on public users' personal computers—38,740 of them and counting. The grid is built to be a "green" and economical computer network, composed of otherwise idle pre-existing infrastructure that lacks the financial and energy requirements of new computers.
Currently, MolecularEvolution.org offers web service for GARLI only. However, the grid system infrastructure is flexible enough to run almost any program.
"In the future, we hope that more scientists and institutions will participate and that more people will use the tools we have developed for their own projects," said Cummings, who also has an appointment in UMD's Institute for Advanced Computer Studies (UMIACS).
More information: * boinc.umiacs.umd.edu/boinc_map.php
* The research paper, "A Gateway for Phylogenetic Analysis Powered by Grid Computing Featuring GARLI 2.0," Adam L. Bazinet, Derrick J. Zwickl, and Michael P. Cummings, was published online April 30, 2014 in Systematic Biology.
Journal information: Systematic Biology
Provided by University of Maryland