January 24, 2014 feature
The ties that bind: Recreating Darwinian ligand evolution in vitro
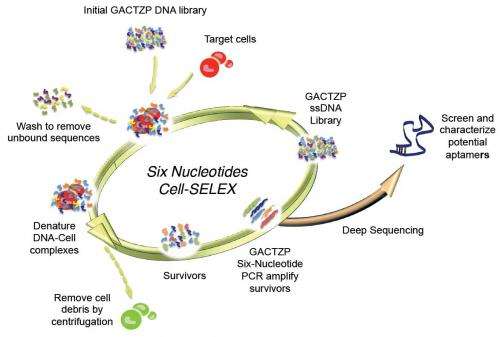
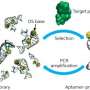
(Phys.org) —A key feature of certain chemicals is their ability to bind to other molecules – a property that emerged through evolution – but current chemical theory lacks the ability to design binders from first principles. To resolve this dilemma, scientists at the University of Florida and the Foundation for Applied Molecular Evolution have recreated these Darwinian processes in vitro (that is, in the laboratory). Their approach extracts nucleic acids as binders from DNA/RNA libraries to evolve through a process called Systematic Evolution of Ligands by Exponential Enrichment, or SELEX. (A ligand is a substance, usually a small molecule, that forms a complex with a biomolecule to serve a biological purpose,) However, natural DNA/RNA has only four nucleotides as building blocks, and so often yields poor binding molecules. By integrating synthetic biology, sequencing tools, and polymerase chain reaction (PCR) amplification, and additional DNA/RNA building blocks from an Artificially Expanded Genetic Information Systems (AEGIS), the scientists successfully demonstrated the first example of SELEX using AEGIS, producing a molecule that binds to cancer cells.
Prof. Steven A. Benner discussed the paper that he, Dr. Kwame Sefah and their co-authors published in Proceedings of the National Academy of Sciences – starting with the main challenges they faced. One such challenge was overcoming the limitation of chemical theory by directly designing binders through in vitro recreation of the Darwinian processes that nature uses to molecular binders. "Current chemical theory is quite able to do broad brush design," Benner tells Phys.org. Indeed, Benner notes, the scientists used current chemical theory to design the additional nucleotides in their AEGIS technology. "However," he continues, "both at the highest level of quantum mechanics as well as the more practical level of molecular dynamics, the assumptions in models of reality are not good enough to allow energy output to be accurate to better than, say, 5 kcal per mole – the difference between success and failure in, as an example, a pharmaceutical."
Conversely, Benner adds, while Darwinism is incapable of doing what he calls big design, it is capable of fine tuning. The whole idea behind this strategy, he explains, is to construct by coarse design a system that is better able to support Darwinism and the laboratory environment. "This publication," he points out, "represents the first case in which this combination has been fully successful." (Benner notes that development of this technology was funded by the Defense Threat Reduction Agency, which consolidates a variety of US Defense Department functions to deal more effectively with the threats posed by nuclear, chemical, or biological weapons.)
A second challenge, says Benner, was demonstrating the first example of SELEX using AEGIS to produce a molecule that binds to cancer cells. "Molecular biological and genetic engineering tools, which have advanced so far over the past half-century, and which have led to several Nobel Prizes, have been advanced for use with natural genetic systems, not our artificially expanded systems. Therefore, we needed to develop analogous molecular biological tools for AEGIS. These included tools to manufacturer the AEGIS components, chemistry to synthesize AEGIS oligonucleotides, enzymes to do AEGIS PCR amplification, procedures to sequence AEGIS DNA, and so on." Incremental research and improvements in technology have allowed AEGIS DNA to be manipulated almost the same level as natural DNA is by classical molecular biology.
To address these challenges, the scientists had to develop chemical pipelines to synthesize AEGIS components on multi-gram scales; develop DNA and RNA polymerase to make DNA and RNA that contain AEGIS components; and create tools to restrict decomposition of DNA containing AEGIS components.
Benner points out that because SELEX, as originally proposed by Larry Gold two decades ago, follows a simple recipe, their findings could possibly generate receptors, ligands, and catalysts having sequence diversities nearer to that displayed by proteins. Specifically, that recipe entails:
1. Synthesizing a library of xNA (xeno-nucleic acid, or synthetic DNA) molecules, typically containing 1011-1014 members.
2. Placing the library in contact with the target in order to separate molecules in the library that bind to the target from molecules that do not. (To select catalysts, xNA libraries are put in an environment where an ability to catalyze a reaction separates them from xNA molecules that cannot.)
3. Survivors are enriched in a pool of xNA molecules that bind to the target (or catalyze a reaction), and then amplified in a polymerase chain reaction (PCR).
4. After a sufficient number of rounds of selection, individual aptamers (oligonucleic acid or peptide molecules that bind to a specific target molecule) are recovered, sequenced, re-synthesized and characterized as molecular species having defined molecular structures.
"As with many new technologies," Benner explains, "SELEX was initially regarded as having high potential, and aptamers were hoped to eventually rival antibodies as binders. This potential has been fulfilled in part. xNA aptamers are known for many targets, including carbohydrates, small molecules, and peptides." In fact, he adds, some aptamers have already entered the clinic – although only after extensive modification of the aptamers originally selected.
"However, as SELEX evolved as a field," Benner continues, "it became clear that the binding diversity and catalytic power of xNA aptamers built from standard nucleotides (G, A, C, and T/U) were not able to match the diversity and power of proteins, including antibodies. At the same time, targets for the best aptamers had a natural propensity to bind to xNA, yielding aptamers with nanomolar dissociation constants." Dissociation constants are a specific type of equilibrium constant that measure the propensity of a larger object to reversibly separate, or dissociate, into smaller components. Ligands having nanomolar (nM) dissociation constants bind more tightly to a particular protein than those with micromolar (μM) dissociation constants. (A molar is a measure of amount-of-substance concentration.)
"For example, Benner illustrates, "aptamers selected to bind to nucleocapsid proteins, reverse transcriptase and HIV integrase had affinities of 2 nM, 0.3-20 nM, and 10-800 nM, respectively. In contrast, aptamers against small molecules were generally selected to have poorer affinities in the micromolar range. For example, dissociation constant values reported for aptamers selected to bind citrulline and arginine ranged from 0.3 to 65 μM, while the values for specific aptamers that bind ATP and xanthine were only 3.3 and 6 μM, respectively."
Benner point out that similar disappointment was also the rule for xNAzymes (xNA molecules that catalyze a preselected reaction). "While many xNAzymes have been reported, the best again performed well only for reactions of nucleic acids themselves, including phosphate hydrolysis, transesterification (the process of exchanging the organic group R″ of an ester with the organic group R′ of an alcohol), xNA ligation, and xNA polymerization. Even here, however, the rates of reactions were not impressive, and certainly not impressive when compared to natural enzymes that have evolved over millions of years to catalyze a specific reaction." He adds that they also did not perform orders of magnitude better than designed protein catalysts or antibody-based catalysts.
"In retrospect," Benner points out, "we might have expected xNA aptamers to not perform as well as proteins, since the latter are built from 20 different units and carry a wide range of chemical functionalities." These include positively-charged nitrogens (on lysine and arginine), anionic groups (aspartate and glutamate), hydrophobic groups (valine, leucine and others), polarizable groups (such as the tryptophan indole and the methionine thioether), catalytic units (including histidine), and metal coordinating groups (cysteine and histidine). In contrast, he adds, standard xNA nucleotides carry little of this functionality.
"Accordingly," Benner continues, "the Benner group and others added functionality to the four standard nucleotides in an attempt to get better performing xNA molecules – and this too has had some success." Examples include:
- Prof. David Perrin and his coworkers1 made DNA where each of the four standard xNA building blocks (GACT) carried a different functional group; applying these to select for DNA catalysts, they saw an improvement in the catalytic power of the emerging DNAzymes (DNA molecules that have the ability to perform a chemical reaction, such as catalytic action)
- SomaLogic modifies the 5-position of pyrimidines with benzyl, naphthyl, tryptamino, and isobutyl groups, inter alia, generating SOMAmers (Slow Off-rate Modified Aptamers); the company reports a set of these that detect ∼800 different human proteins in blood volumes as small as 15 microliters (μL), with affinities in the 0.1 picomolars (pM) - 1 μM range
Benner also tells Phys.org that AEGIS–SELEX would be improved by the following investigations being developed in their laboratories:
- Strains of E. coli cells that can propagate plasmids containing GACTZP DNA (a form of xNA), which he notes is the core of their current activities
- A technology for directly sequencing single molecule Z:P pairs
- Polymerases with still higher levels of fidelity and efficiency
Moving forward, Benner tells Phys.org, the scientists are developing E. coli strains that can propagate plasmids containing GACTZP DNA, and are improving sequencing. "We also hope to also develop the chemistry and enzymology of the xNA S:B pair, as well as the xNA K:X and V:J pairs, in this order."
Benner notes that there are other areas of research that might benefit from their study. "Nearly everything in medicine involves the binding of molecules to other molecules," he tells Phys.org. "This is true in the research laboratory, in clinical diagnostics, and even within patients. Medicine needs drugs that bind to therapeutic targets to cure diseases; diagnostics needs molecules that bind to markers to indicate the presence of the diseased state; and researchers need molecules that bind to functional molecules to track their movement within cells." For this reason, he says, binding to and between targets is key to essentially all work done under the auspices of the National Institutes of Health (NIH), even when that binding is not the explicit focus of that work.
"Accordingly," Benner concludes, "biomedical researchers, diagnosticians and clinicians have long sought a technology that delivers binding molecules and catalysts on demand. In this vision, the technology would deliver binders quickly, reliably and inexpensively, to any researcher who has in hand some of the target. AEGIS-SELEX will be, we hope, this technology."
More information: In vitro selection with artificial expanded genetic information systems, PNAS Published online before print December 30, 2013, doi:10.1073/pnas.1311778111
Related:
1Toward the Combinatorial Selection of Chemically Modified DNAzyme RNase A Mimics Active Against all-RNA Substrates, ACS Combinatorial Science 2013 15 (4), 174-182, doi:10.1021/co3001378
Journal information: Proceedings of the National Academy of Sciences
© 2014 Phys.org. All rights reserved.