Squeezing more production out of bacteria
Engineering microorganisms to manufacture chemicals is not a new idea: In the 1980s, scientists figured out how to get bacteria to produce insulin, which diabetics need to control their disease. This technique, known as metabolic engineering, involves inserting many copies of the gene for a desired compound into bacteria or yeast cells. Scientists have also used this approach to turn bacteria into tiny factories that can generate biofuels such as ethanol, as well as plastics.
MIT chemical engineering professor Kristala Jones Prather and colleagues are now taking genetic manipulation a step further. By tinkering with the genes before inserting them into bacteria, they can manipulate each step of a synthetic reaction inside a cell. That strategy, known as protein engineering, lets researchers use bacteria to make new products, or to boost production of naturally occurring compounds.
By combining protein engineering with traditional metabolic engineering, Prather and her colleagues recently induced E. coli to synthesize a chemical normally secreted by plants, at a rate 2,600 times that of the natural reaction, and about four times greater than using traditional metabolic engineering alone. This approach could be applied to any kind of synthetic pathway inside a cell, including those that produce pharmaceutical compounds or biofuels, says Prather.
In a study recently published in the Proceedings of the National Academy of Sciences, Prather and postdoctoral associates Effendi Leonard and Ajikumar Parayil focused on a biological pathway that produces a class of compounds known as terpenoids. There are at least 60,000 known terpenoids, many of which are produced by plants and contribute to the distinctive flavors of cinnamon, cloves and ginger.
In plants, terpenoids serve as chemoattractants, enticing insects to pollinate them, or repelling predators. Of more interest to humans, terpenoids can be converted to ginkgolides, naturally produced by the ginkgo biloba plant. Ginkgolides have been associated with memory enhancement, though their effectiveness has not been proven. They are also being investigated for treatment of multiple sclerosis and vitiligo, a skin disorder.
Obtaining large quantities of terpenoids or gingkolides is difficult because plants don’t produce them in large quantities, and there is no way to synthesize them in a laboratory.
Targeting enzymes
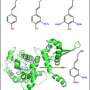
The MIT team, which also included professors Gregory Stephanopoulos and Bruce Tidor, focused on a reaction pathway whose end product, levopimaradiene, is a terpenoid that can be converted to ginkgolides. Bacteria don’t normally produce levopimaradiene, so the researchers added genes for two plant enzymes that can convert chemicals found in bacteria into levopimaradiene.
However, those enzymes don’t yield enough of the desired product, so the researchers started trying to redesign the enzymes to make them more efficient. They started with an enzyme called LPS, which catalyzes the last step of the process, converting a compound called GGPP to levopimaradiene. LPS can also convert GGPP to three other products, so the researchers tried to create a new version that would generate levopimaradiene exclusively.
To do this, they identified a stretch of 15 amino acids in the enzyme where altering the sequence should have the greatest effect, and tested about 120 proteins with different sequences. Using this very targeted approach, they found an enzyme variation that produces 97 percent levopimaradiene (compared to 85 percent for the original enzyme), and is about five times faster than the original.
Though highly successful, this approach only works when the researchers have a good structural model of the original protein, says Prather.
Next, the researchers tackled the second-to-last step of the process, which is the conversion of two compounds naturally found in bacteria to GGPP by an enzyme. For this step, they used a more traditional protein-engineering approach, creating about 1 million variants of the original protein, then testing their enzymatic efficiency. This less targeted approach usually works only if there is a way to rapidly test large numbers of proteins, which there was in this case. Their final enzyme was twice as fast as the original.
The researchers also gained a 600-fold boost in productivity by only using traditional metabolic engineering, adding extra copies of three genes for enzymes involved in earlier stages of terpenoid synthesis. Combining protein engineering resulted in an overall levopimaradiene yield 2,600 times greater than that of the unengineered enzymes. In adding extra copies, the researchers have to be careful not to add too many, which can overwhelm the bacteria and actually slow down production.
Next, Prather plans to apply this strategy to other pathways, including one that produces hydroxy acids, which are useful for drug manufacturing. She has patented the mutant bacterial strains produced in the PNAS study, but is not planning to commercialize the process.
Claudia Schmidt-Dannert, associate professor of biochemistry at the University of Minnesota, says she believes this approach should prove useful in engineering many different types of metabolic pathways. “Just making these little improvements in bottleneck enzymes can have a big impact on the overall production of the pathway,” she says.
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.
Provided by Massachusetts Institute of Technology