At the crossroads of chromosomes: Study reveals structure of cell division's key molecule
(PhysOrg.com) -- On average, one hundred billion cells in the human body divide over the course of a day. Most of the time the body gets it right but sometimes, problems in cell replication can lead to abnormalities in chromosomes resulting in many types of disorders, from cancer to Down Syndrome.
Now, researchers at the University of Pennsylvania’s School of Medicine have defined the structure of a key molecule that plays a central role in how DNA is duplicated and then moved correctly and equally into two daughter cells to produce two exact copies of the mother cell. Without this molecule, entire chromosomes could be lost during cell division.
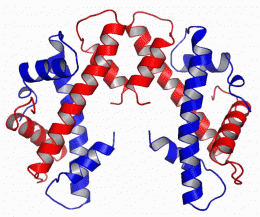
Ben Black, PhD, assistant professor of Biochemistry and Biophysics, and Nikolina Sekulic, PhD, a postdoctoral fellow in the Black lab, report in the September 16 issue of Nature the structure of the CENP-A molecule, which defines a part of the chromosome called the centromere. This is a constricted area to which specialized molecules called spindle fibers attach that help pull daughter cells apart during cell division.
“Our work gives us the first high-resolution view of the molecules that control genetic inheritance at cell division,” says Black. “This is a big step forward in a puzzle that biologists have been chipping away at for over 150 years.”
Investigators have known for the last 15 years that part of cell division is controlled by epigenetic processes, the series of actions that affect the protein spools around which DNA is tightly bound, rather than encoded in the DNA sequence itself. Those spools are built of histone proteins, and chemical changes to these spool proteins can either loosen or tighten their interaction with DNA. Epigenetics alter the readout of the genetic code, in some cases ramping a gene’s expression up or down. In the case of the centromere, it marks the site where spindle fibers attach independently of the underlying DNA sequence. CENP-A has been suspected to be the key epigenetic marker protein.
However, what hasn’t been known is how CENP-A epigenetically marks the centromere to direct inheritance. The Black team found the structural features that confer CENP-A the ability to mark centromere location on each chromosome. This is important because without CENP-A or the centromere mark it creates, the entire chromosome—and all of the genes it houses—are lost at cell division.
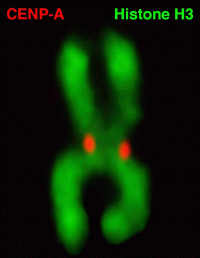
In this study, Black solved CENP-A’s structure to determine how it specifically marks the centromere on each chromosome and surmise from that how the epigenetic mark is copied correctly in each cell division. They found that CENP-A changes the shape of the nucleosome of which it’s a part, also making it more rigid than other nucleosomes without CENP-A. The nucleosome is the combination of DNA wound around a histone protein core --the DNA thread wrapped around the histone spool. The CENP-A nucleosome is copied several times to create a unique epigenetic area, different from the rest of the chromosome. CENP-A replaces histone H3 in the nucleosomes located at the centromere.
This CENP-A centromere identifier attracts other proteins, and in cell division builds a massive structure, the kinetochore, for pulling the duplicated chromosomes apart during cell division.
Besides the major advance in the understanding of the molecules driving human inheritance, this work also brings about the exciting prospect that the key epigenetic components are now in hand to engineer clinically useful artificial chromosomes that will be inherited alongside our own natural chromosomes—and with the same high fidelity, says Black.
Provided by University of Pennsylvania School of Medicine















