Key enzyme discovered to be master regulator in protein-protein reactions
Protein phosphorylation is a process by which proteins are flipped from one activation state to another. It is a crucial function for most living beings, since phosphorylation controls nearly every cellular process, including metabolism, gene transcription, cell-cycle progression, cytoskeletal rearrangement and cell movement.
Due to its importance in biology, scientists have wanted to learn more about protein phosphorylation and how proteins know when and how to become phosphorylated or dephosphorylated. Think of it like choosing the right dance partner who knows your moves so intimately that the choreography is seamless. Biologists have learned that interactions by kinases (enzymes that add a phosphate to a protein) are highly regulated. Each of the 428 human serine/threonine kinases interact only with certain substrate proteins, and they pick their "partners" unfailingly. But for the reverse reaction, called dephosphorylation (removing a phosphate from a protein), only about 40 phosphatases are available to interact with all substrate proteins. In fact, just one of them, protein phosphatase 1 (PP1), is believed to be responsible for up to 65 percent of dephosphorylation reactions.
The question then is how PP1, a generalist, knows which substrate proteins to interact with. New research by Wolfgang Peti, the Manning Assistant Professor of Medical Science and assistant professor of chemistry, reported in a paper published online in Nature Structural & Molecular Biology, helps to answer that question. Peti and colleagues at Brown and Yale University have discovered that PP1 "chooses" proteins in dephosphorylation reactions based on which of its binding sites is available for the interaction to occur. The finding is important, because erroneous PP1 regulation can cause numerous diseases, including cancer (chromatin remodeling), diabetes (glycogen) and Parkinson's (LTP).
"There are thousands of (peer-reviewed) papers out there, but nobody understood how PP1 is regulated," said Peti. "It is in fact a master regulator. We have identified now how it works."
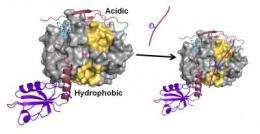
To obtain their results, the team showed for the first time how PP1 is bound to a regulator protein, spinophilin. Using nuclear magnetic resonance spectroscopy and X-ray crystallography, the researchers examined the spinophilin-PP1 complex's structure. The scientists saw that spinophilin had attached itself to one of three substrate binding sites on PP1, called the C-terminal substrate binding groove. Examined at the atomic scale, it appears as if spinophilin is a many-tentacled beast that has woven itself into the C-terminal substrate binding groove,
effectively blocking any substrate requiring this groove from interacting with PP1.
That leaves only two other binding sites, the acidic and hydrophobic substrate binding grooves. "Any substrate that needs the C-terminal is out of the game," Peti said.
By narrowing the binding sites from three to two, PP1 is in effect becoming more selective, Peti noted. "What that means is PP1 is equally tightly controlled as kinases are," he said.
"Now we know how PP1 is regulated," Peti added. "What simply happens is we don't create more enzymes. We create more protein complexes (holoenzymes) that increase the specificity of PP1."
Provided by Brown University