Scientists develop universal DNA reader to advance faster, cheaper sequencing efforts
Arizona State University scientists have come up with a new twist in their efforts to develop a faster and cheaper way to read the DNA genetic code. They have developed the first, versatile DNA reader that can discriminate between DNA's four core chemical components⎯the key to unlocking the vital code behind human heredity and health.
Led by ASU Regents' Professor Stuart Lindsay, director of the Biodesign Institute's Center for Single Molecule Biophysics, the ASU team is one of a handful that has received stimulus funds for a National Human Genome Research Initiative, part of the National Institutes of Health, to make DNA genome sequencing as widespread as a routine medical checkup.
The broad goal of this "$1000 genome" initiative is to develop a next-generation DNA sequencing technology to usher in the age of personalized medicine, where knowledge of an individual's complete, 3 billion-long code of DNA information, or genome, will allow for a more tailored approach to disease diagnosis and treatment. With current technologies taking almost a year to complete at a cost of several hundreds of thousands of dollars, less than 20 individuals on the planet have had their whole genomes sequenced to date.
To make their research dream a reality, Lindsay's team has envisioned building a tiny, nanoscale DNA reader that could work like a supermarket checkout scanner, distinguishing between the four chemical letters of the DNA genetic code, abbreviated by A, G, C, and T, as they rapidly pass by the reader.
To do so, they needed to develop the nanotechnology equivalent of threading the eye of a needle. In this case, the DNA would be the thread that could be recognized as it moved past the reader 'eye.' During the past few years, Lindsay's team has made steady progress, and first demonstrated the ability to read individual DNA sequences in 2008—but this approach was limited because they had to use four separate readers to recognize each of the DNA bases. More recently, they demonstrated the ability to thread DNA sequences through the narrow hole of a fundamental building block of nanotechnology, the carbon nanotube.
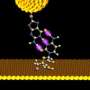
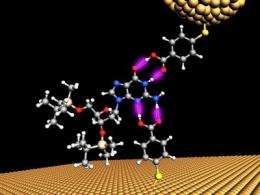
Lindsay's team relies on the eyes of nanotechnology, scanning tunneling- (STM) and atomic force- (ATM) microscopes, to make their measurements. The microscopes have a delicate electrode tip that is held very close to the DNA sample.
In their latest innovation, Lindsay's team made two electrodes, one on the end of microscope probe, and another on the surface, that had their tiny ends chemically modified to attract and catch the DNA between a gap like a pair of chemical tweezers. The gap between these functionalized electrodes had to be adjusted to find the chemical bonding sweet spot, so that when a single chemical base of DNA passed through a tiny, 2.5 nanometer gap between two gold electrodes, it momentarily sticks to the electrodes and a small increase in the current is detected. Any smaller, and the molecules would be able to bind in many configurations, confusing the readout, any bigger and smaller bases would not be detected.
"What we did was to narrow the number of types of bound configurations to just one per DNA base," said Lindsay. "The beauty of the approach is that all the four bases just fit the 2.5 nanometer gap, so it is one size fits all, but only just so!"
At this scale, which is just a few atomic diameters wide, quantum phenomena are at play where the electrons can actually leak from one electrode to the other, tunneling through the DNA bases in the process.
Each of the chemical bases of the DNA genetic code, abbreviated A, C, T or G, gives a unique electrical signature as they pass between the gap in the electrodes. By trial and error, and a bit of serendipity, they discovered that just a single chemical modification to both electrodes could distinguish between all 4 DNA bases.
"We've now made a generic DNA sequence reader and are the first group to report the detection of all 4 DNA bases in one tunnel gap," said Lindsay. "Also, the control experiments show that there is a certain (poor) level of discrimination with even bare electrodes (the control experiments) and this is in itself, a first too."
"We were quite surprised about binding to bare electrodes because, like many physicists, we had always assumed that the bases would just tumble through. But actually, any surface chemist will tell you that the bases have weak chemical interactions with metal surfaces."
Next, Lindsay's group is hard at work trying to adapt the reader to work in water-based solutions, a critically practical step for DNA sequencing applications. Also, the team would like to combine the reader capabilities with the carbon nanotube technology to work on reading short stretches of DNA.
If the process can be perfected, DNA sequencing could be performed much faster than current technology, and at a fraction of the cost. Only then will the promise of personalized medicine reach a mass audience.
More information: The Nano Letters research article can be accessed online at URL: pubs.acs.org/doi/pdfplus/10.1021/nl1001185
Provided by Arizona State University