Researchers capture bacterial infection on film (w/ Video)
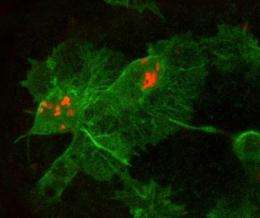
(PhysOrg.com) -- Researchers have developed a new technique that allows them to make a movie of bacteria infecting their living host.
Whilst most studies of bacterial infection are done after the death of the infected organism, this system developed by scientists at the University of Bath and University of Exeter is the first to follow the progress of infection in real-time with living organisms.
The researchers used developing fruit fly embryos as a model organism, injecting fluorescently tagged bacteria into the embryos and observing their interaction with the insect's immune system using time-lapse confocal microscopy.
The researchers can also tag individual bacterial proteins to follow their movement and determine their specific roles in the infection process.
The scientists are hoping to use this system in the future with human pathogens such as Listeria and Trypanosomes. By observing how these bacteria interact with the immune system, researchers will gain a better understanding of how they cause an infection and could eventually lead to better antibacterial treatments.
Dr Will Wood, Research Fellow in the Department of Biology & Biochemistry at the University of Bath, explained: "Cells often behave very differently once they have been taken out of their natural environment and cultured in a petri dish.
"In the body, immune surveillance cells such as hemocytes (or macrophages in vertebrates) are exposed to a battery of signals from different sources. The cells integrate these signals and react to them accordingly.
"Once these cells are removed from this complex environment and cultured in a petri dish these signals are lost. Therefore it is really important to study whole organisms to fully understand how bacteria interact with their host."
Dr Nick Waterfield, co-author on the study and Research Officer at the University of Bath, said: "To be able to film the microscopic battle between single bacterial cells and immune cells in a whole animal and in real time is astounding.
"It will ultimately allow us to properly understand the dynamic nature of the infection process."
Professor Richard Ffrench-Constant, Professor of Molecular Natural History at the University of Exeter, added: "For the first time this allows us to actually examine infection in real time in a real animal - it's a major advance!"
The study has been published in PLoS Pathogens.
Source: University of Bath (news : web)