The future of personalized cancer treatment: An entirely new direction for RNAi delivery
In technology that promises to one day allow drug delivery to be tailored to an individual patient and a particular cancer tumor, researchers at the University of California, San Diego School of Medicine, have developed an efficient system for delivering siRNA into primary cells. The work will be published in the May 17 in the advance on-line edition of Nature Biotechnology.
"RNAi has an unbelievable potential to manage cancer and treat it," said Steven Dowdy, PhD, Howard Hughes Medical Institute Investigator and professor of cellular and molecular medicine at UC San Diego School of Medicine. "While there's still a long way to go, we have successfully developed a technology that allows for siRNA drug delivery into the entire population of cells, both primary and tumor-causing, without being toxic to the cells."
For many years, Dowdy has studied the cancer therapy potential of RNA inhibition which can be used to silence genes through short interfering, double-stranded RNA fragments called siRNAs. But delivery of siRNAs has proven difficult due to their size and negative electrical charge - which prohibits them from readily entering cells.
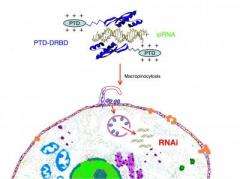
A small section of protein called a peptide transduction domain (PTD) has the ability to permeate cell membranes. Dowdy and colleagues saw the potential for PTDs as a delivery mechanism for getting siRNAs into cancer cells. He and his team had previously generated more than 50 "fusion proteins" using PTDs linked to tumor-suppressor proteins.
"Simply adding the siRNAs to a PTD didn't work, because siRNAs are highly negatively charged, while PTDs are positively charged, which results in aggregation with no cellular delivery," Dowdy explained. The team solved the problem by making a PTD fusion protein with a double-stranded RNA-binding domain, termed PTD-DRBD, which masks the siRNA's negative charge. This allows the resultant fusion protein to enter the cell and deliver the siRNA into the cytoplasm where it specifically targets mRNAs from cancer-promoting genes and silences them.
To determine the ability of this PTD-DRBD fusion protein to deliver siRNA, the researchers generated a human lung cancer reporter cell line. Using green and fluorescent protein and analyzing the cells using flow cytometry analysis, they were able to determine the magnitude of RNA inhibitory response and the percentage of cells undergoing this response. They found that the entire cellular population underwent a maximum RNAi response. Similar results were obtained in primary cells and cancer cell lines.
"We were subsequently able to introduce gene silencing proteins into a large percentage of various cell types, including T cells, endothelial cells and human embryonic stem cells," said Dowdy. "Importantly, we observed no toxicity to the cells or innate immune responses, and a minimal number of transcriptional off-target changes."
These RNAi methods can be continually tweaked to combat new mutations - a way to overcome a major problem associated with current cancer therapies. "Such therapies can't be used a second time if a cancer tumor returns, because the tumor has mutated the target gene to avoid the drug binding," said Dowdy. "But since the synthetic siRNA is designed to bind to a single mutation and only that mutation on the genome, it can be easily and rapidly changed while maintaining the delivery system - the PTD-DRBD fusion protein."
"Cancer is a complex, genetic disease that is different in every patient," Dowdy added. "This is still in early stages, but I believe the siRNA-induced RNAi approach to personalized cancer treatment is the only thing on the table."
Source: University of California - San Diego (news : web)
















