Proteins by design: Biochemists create new protein from scratch
(PhysOrg.com) -- No doubt proteins are complex. Most are "large" and full of interdependent branches, pockets and bends in their final folded structure. This complexity frustrates biochemists and protein engineers seeking to understand protein structure and function in order to reproduce or create new uses for these natural molecules to fight diseases or for use in industry.
Using design and engineering principles learned from nature, a team of biochemists from the University of Pennsylvania School of Medicine have built - from scratch - a completely new type of protein. This protein can transport oxygen, akin to human neuroglobin, a molecule that carries oxygen in the brain and peripheral nervous system. Some day this approach could be used to make artificial blood for use on the battle field or by emergency-care professionals. Their findings appear in the most recent issue of Nature.
"This is quite a different way of making novel proteins than the rest of the world," says senior author P. Leslie Dutton, PhD, Eldridge Reeves Johnson Professor of Biochemistry and Biophysics. "We've created an unusually simple and relatively small protein that has a function, which is to carry oxygen. No one else has ever done this before."
"Our aim is to design new proteins from principles we discover studying natural proteins," explains co-author Christopher C. Moser, PhD, Associate Director of the Johnson Foundation at Penn. "For example, we found that natural proteins are complex and fragile and when we make new proteins we want them to be simple and robust. That's why we're not re-engineering a natural protein, but making one from scratch."
Currently, protein engineers take an existing biochemical scaffold from nature and tweak it a bit structurally to make it do something else. "This research demonstrates how we used a set of simple design principles, which challenge the kind of approaches that have been used to date in reproducing natural protein functions," says Dutton.
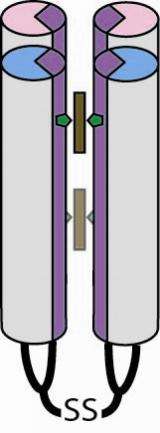
To build their protein, the Penn team started with just three amino acids, which code for a helix-shaped column. From this, they assembled a four-column bundle with loops that resembles a simple candelabra. They added a heme, a chemical group that contains an iron atom, to bind oxygen molecules. They also added another amino acid called glutamate to add strain to the candelabra to help the columns open up to capture the oxygen. Since heme and oxygen degrade in water, the researchers also designed the exteriors of the columns to repel water to protect the oxygen payload inside.
Initially, the team used a synthesizer -- a robot that chemically sticks amino acids together in a desired sequence -- to make the helix-shaped columns. "We do the first reactions with the robot to figure out the sequence of amino acids that we want," says Moser. When they are satisfied with the sequence, they use the bacterium E. coli as a biological host to make the full protein.
The team used chemical tests to confirm that their protein did indeed capture oxygen. When the oxygen did bind to the iron heme molecule in the artificial protein, the solution in which the reaction took place changed color from dark red to scarlet, a color signature almost identical to natural neuroglobin.
"This exercise is like making a bus," says Dutton. "First you need an engine and we've produced an engine. Now we can add other things on to it. Using the bound oxygen to do chemistry will be like adding the wheels. Our approach to building a simple protein from scratch allows us to add on, without getting more and more complicated."
Source: University of Pennsylvania School of Medicine (news : web)