Genome editing: Efficient CRISPR experiments in mouse cells
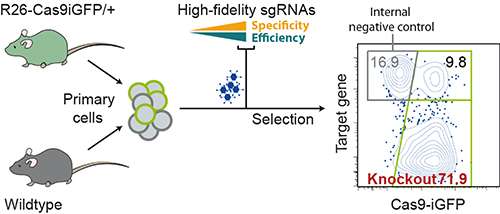
In order to use the CRISPR-Cas9 system to cut genes, researchers must design an RNA sequence that matches the DNA of the target gene. Most genes have hundreds of such sequences, with varying activity and uniqueness in the genome. The search for the best sequences is therefore hardly achievable by hand. The new "CrispRGold" program helps scientists to identify the most effective and specific RNA sequences. It has been devised by a group of researchers headed by Prof. Klaus Rajewsky of the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), and is now described in the journal PNAS. The team has also developed a new mouse model that already carries the Cas9 protein. Combining this mouse model with the reliable RNA sequences allowed an efficient inactivation of genes in primary cells. This has enabled the researchers to discover new genes involved in the regulation of immune cells.
For many molecular biologists, the discovery of the CRISPR-Cas9 system marked a new milestone in research: finally, genomic DNA can be cut with high efficiency and precision, enabling genes to be disabled, modified or re-introduced.
This requires little more than a snippet of RNA from the genetic material, which takes the Cas9 protein scissors to the point in the DNA that is to be cut. This RNA snippet, known as sgRNA (single guide RNA), contains a sequence of 20 RNA-letters complementary to the genomic target site that scientists have hitherto had to select laboriously by hand or with a variety of online tools. In some cases, it was then uncertain whether the sgRNA would take the Cas9 gene scissors to the right place or to a similar but unwanted place in the genome and whether the sgRNA efficiency was high.
The new "CrispRGold" program written by PhD student Robin Graf from the MDC research group headed by Prof. Klaus Rajewsky makes it significantly easier to disable specific genes.
The program searches a defined DNA target sequence in order to identify the best place for the cut and suggests an sgRNA sequence that is unique in the genetic material and hence delivers the Cas9 protein only to the required point. Cas9 can then snip the gene so that it ceases to function. The algorithm is based on experimental data as well as the uniqueness and other properties of the sequences.
With his colleague Dr. Van Trung Chu, Graf tested the system on certain white blood cells in the mouse, the B cells. These cells cannot be cultivated for any length of time, because they do not survive long outside their natural environment. Genes must therefore be deactivated quickly and in as many cells as possible in order to study their function. Chu achieved this by breeding a genetically modified line of mice that produces large yet well-tolerated quantities of the Cas9 gene scissors. The researchers then isolated the B cells from these mice and delivered sgRNAs specific for individual genes to these cells. With high reproducibility, the sgRNAs designed with CrispRGold destroyed the target genes in on average 80 percent of the cells - "an excellent rate", says Graf. "High efficiency and a low error rate are absolutely essential in low-throughput experiments of this sort."
The researchers used their new method to identify a number of previously unknown genes that are involved in B-cell development. The CrispRGold program will now be made available online so that it can be used by scientists worldwide: "The program can easily be used for other types of cell from a wide range of organisms. It could also be relevant to clinical applications - it treats sequence uniqueness as a high priority and thus minimises the risk of potentially unwanted gene modifications, which must be avoided at all costs in gene therapy," says Graf. CrispRGold is expected to be available at http://crisprgold.mdc-berlin.de from November 2016.
More information: Van Trung Chu et al, Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line, Proceedings of the National Academy of Sciences (2016). DOI: 10.1073/pnas.1613884113
Journal information: Proceedings of the National Academy of Sciences
Provided by Max Delbrück Center for Molecular Medicine