Biochemists advancing research into autism, Alzheimer's disease and Down syndrome

Biochemists at The University of Texas at Arlington are mapping the catalytic processes of sulfur-oxidizing enzymes to improve understanding of the chemical imbalances found in patients with autism, Alzheimer's disease and Down syndrome.
"Little is known about how sulfur-oxidizing enzymes work, or how or why autistic, Alzheimer and Down syndrome patients demonstrate abnormal sulfur metabolism," said Brad Pierce, UTA assistant professor of biochemistry and principal investigator on the project.
"Our work is to retro-engineer the sulfur oxidation process and map out the chemical mechanism of a key enzyme - cysteine dioxygenase - in both mammals and bacteria, to provide the necessary framework to develop effective therapies and drugs for these different disease states."
Insights into the differential behavior of this enzyme in bacteria could also open up opportunities to stamp out "superbugs" by providing an alternate means to disrupt bacterial metabolism without adversely affecting the patient, Pierce said.
The work is supported by a three-year, $333,810 National Institutes of Health grant through the agency's Academic Research Enhancement Award Program, which aims to strengthen the research environment at eligible institutions and also expose undergraduate and graduate students to research and careers in science, technology, engineering and mathematics.
Morteza Khaledi, dean of the UTA College of Science, underlined the importance of Pierce's grant in enhancing the University's commitment to advancing health and the human condition, as outlined in the Strategic Plan 2020: Bold Solutions | Global Impact.
"Dr. Pierce's research will provide the essential basic scientific background needed to develop therapies for critical conditions that are currently not understood," Khaledi said. "This work is at the heart of the positive impact that a modern, research university can have on society."
"By providing opportunities to undergraduate and graduate students to participate in this research, we are also strengthening our position as a source of well-prepared scientific resources and advancing towards Tier 1 status," he added.
In the current project, Pierce's team will use rapid-mix, freeze-quench techniques to 'trap' and analyze the chemical reactions at millisecond intervals. Comparisons can then be made between the mechanical processes of the enzymes in mammals and bacteria.
This research builds on Pierce's prior National Science Foundation grant to study the circumstances under which cysteine dioxygenase produces highly toxic side effects called reactive oxygen species. Those effects have been linked to numerous age-onset human diseases like cancer, stroke, arthritis, heart attacks, Parkinson's disease, cataracts and many others.
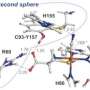
In a study published in the December 2013 issue of Biochemistry, Pierce's team outlined how mutations outside the active site environment or "outer coordination sphere" of the enzyme have profound influence on the release of reactive oxygen species. Prior research had focused on the active site inner coordination sphere of these enzymes, where the metal molecule is located.
Pierce joined the UTA College of Science in 2008 following positions as an NIH postdoctoral researcher at the University of Wisconsin and as a research associate with a California pharmaceutical company. He earned his doctorate in chemistry from Carnegie Mellon University.
At UTA, Pierce has been honored with a President's Award for Excellence in Teaching. His research group looks at fundamental life processes outside the traditional sphere of biochemistry and employs modern biophysical and bioinorganic techniques to investigate enzyme function and regulation.
Provided by University of Texas at Arlington