February 17, 2015 report
Biologist describes optimized cellular replication as a systems engineering problem
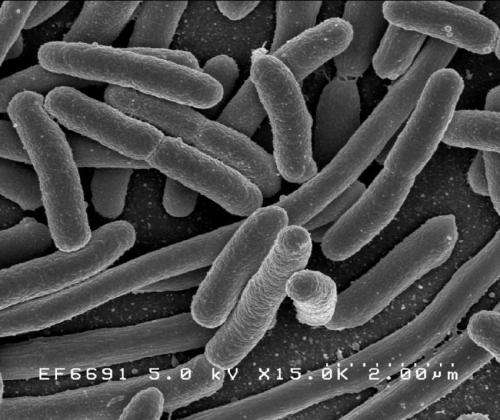
(Phys.org)—Biologist Rami Pugatch of Princeton's Simons Center for Systems Biology has characterized the self-replication process of Escherichia coli according to a scheduling policy model derived from industrial processes. The paper, titled "Greedy scheduling of cellular self-replication leads to optimal doubling times with a log-Frechet distribution," appears in the Proceedings of the National Academy of Sciences.
The paper describes the problem of task scheduling in cellular replication processes and ultimately produces a mathematical distribution that characterizes an optimal replication strategy for E. coli cells. The scope of Pugatch's work encompasses individual cellular processes, algorithmic descriptions of optimized replication, systems engineering concepts, and even the history of the concept of the self-replicating factory.
Interestingly, mathematician John von Neumann's thought experiment regarding self-replicating machines, presented in 1948, predicted the cellular systems explained in the paper even before the discovery of the molecular structure of DNA. His model, which he called a "universal constructor," described a factory machine that reads instructions and translates them into the assembly of every other machine in the factory, including itself, provided that all of the necessary substrates are available.
Such a factory is called "non-trivial" if it includes a universal constructor as a component. The duplicative process is not considered to be complete until a copy of the instructions is provided. Instead of directing their own replication, the instructions are instead duplicated from a template by a separate dedicated machine that is not triggered until the completion of the factory replication phase. This is closely analogous to actual cellular processes.
The scheduling problem
However, in its scheduling of processes, this model assumes serial dynamics that do not correspond with the more complex parallel processes of cellular mechanics. Pugatch describes a "scheduling problem" in which cells must optimize the allocation of resources for self-replication in order to thrive in a competitive environment. If the number of processing units available is smaller than the number of cellular production tasks that require them, it is computationally difficult to find an optimal schedule.
To complete a biosynthetic task, associated catalysts and production materials must be allocated. Assuming that there is an abundant supply of input materials, the problem lies in assigning tasks to processing units such that the completion time of the entire task is minimized. "In the noisy cellular milieu, we cannot expect scheduling algorithms to be precise," Pugatch writes. "Instead, we ask what is expected from a randomized algorithm."
He finds that the result is "greedy scheduling"—in a condition of excess catalysts, demand for them is easily met as long as they are never idle if there is an available task. Thus, cells favor excess, though it comes at the cost of wasteful free energy, because the ready availability of catalysts results in fast replication.
The replicative buffer
His study of E. coli bacteria growing exponentially in a stationary medium reveals a measured distribution of doubling times that corresponds with the predicted doubling times of an optimally scheduled von Neumann self-replicating factory. To make his analysis, Pugatch employs a systems engineering project graph to define the "replicative buffer"—that is, the number of basic self-replicating units within a cell.
"A replicative buffer greater or equal to 1 allows a large class of random scheduling algorithms known as list algorithms to obtain optimal completion times," he notes. The distribution of optimal completion times is derived via a special type of project graph representing balanced production.
Closure and essentiality
"Closure" and "essentiality" are two unique features of self-replicating factories, including cells. Closure occurs when all of the processing units have converted raw materials into the final product and each processing unit is present in duplicate.
Examples of cellular processing units include metabolic enzymes, RNA and DNA polymerases, and ribosomes. Pugatch notes, "Like processing units in a factory, [these] are required for a production task (biosynthesis), consume input materials and free energy, are not consumed during the process, yet are essential for its successful completion."
The paper illustrates, as an example of closure and essentiality, the production of proteins by ribosomes. The ribosomes are themselves built from ribosomal proteins and ribosomal RNA. To synthesize proteins, ribosomes require a pool of processing units—in this case, charged transfer RNA and a family of auxiliary proteins. Additionally, they need the catalysts for tRNA charging. All elements are considered "essential," as they are required for the production of members of another group of elements. All of the production elements are either synthesized by cellular mechanisms or imported from outside the cell by metabolic processes.
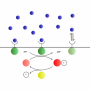
Pugatch's coarse-grained models of individual cellular processes are essentially identical to the von Neumann model, and characterize different temporal organizations of self-replication. By mapping the interactions between the sets of components and the catalytic reactions, Pugatch produces a reaction graph that allows analysis via the project evaluation and review technique (PERT), which is commonly used in systems engineering for scheduling important events in a project.
This analysis reveals that the optimal project completion time can be derived by finding the duration of the longest start-to-termination path among cell processes; this is known as the "critical path." The other, shorter paths have "slack," describing a duration gap between their completion times and that of the critical path. This allows more flexibility in the scheduling of many cellular processes.
"When fast completion time is a desirable goal, one can alleviate the constraints that cause a given critical path to be dominant, possibly creating a new (faster) critical path. Iteratively repeating this constraint elimination process until convergence results in a maximally balanced project," Pugatch writes. The PERT analysis produced a periodic structure of self-replication that Pugatch found has a universal shape—a log-Frechet distribution.
More information: "Greedy scheduling of cellular self-replication leads to optimal doubling times with a log-Frechet distribution," Rami Pugatch. PNAS 2015 ; published ahead of print February 9, 2015, DOI: 10.1073/pnas.1418738112
Abstract
Bacterial self-replication is a complex process composed of many de novo synthesis steps catalyzed by a myriad of molecular processing units, e.g., the transcription–translation machinery, metabolic enzymes, and the replisome. Successful completion of all production tasks requires a schedule—a temporal assignment of each of the production tasks to its respective processing units that respects ordering and resource constraints. Most intracellular growth processes are well characterized. However, the manner in which they are coordinated under the control of a scheduling policy is not well understood. When fast replication is favored, a schedule that minimizes the completion time is desirable. However, if resources are scarce, it is typically computationally hard to find such a schedule, in the worst case. Here, we show that optimal scheduling naturally emerges in cellular self-replication. Optimal doubling time is obtained by maintaining a sufficiently large inventory of intermediate metabolites and processing units required for self-replication and additionally requiring that these processing units be "greedy," i.e., not idle if they can perform a production task. We calculate the distribution of doubling times of such optimally scheduled self-replicating factories, and find it has a universal form—log-Frechet, not sensitive to many microscopic details. Analyzing two recent datasets of Escherichia coli growing in a stationary medium, we find excellent agreement between the observed doubling-time distribution and the predicted universal distribution, suggesting E. coli is optimally scheduling its replication. Greedy scheduling appears as a simple generic route to optimal scheduling when speed is the optimization criterion. Other criteria such as efficiency require more elaborate scheduling policies and tighter regulation.
Journal information: Proceedings of the National Academy of Sciences
© 2015 Phys.org