Illuminating the dark side of the genome

Almost 50 percent of our genome is made up of highly repetitive DNA, which makes it very difficult to be analysed. In fact, repeats are discarded in most genome-wide studies and thus, insights into this part of the genome remained limited. Scientists from the Max Planck Institute of Immunobiology and Epigenetics (MPI-IE) in Freiburg now succeeded in examining this dark side of the genome. Their analyses revealed that repeat-associated heterochromatin is essential to repress retrotransposons and thereby protects the genomic integrity of stem cells. This work opens the way for future genome-wide analyses of repetitive regions in the genome and is in line with newly emerging functions for heterochromatin.
Only 1 % of the human genome contains coding information, while the remaining 99 % harbour non-coding and repetitive DNA. DNA and its packaging proteins (histones) create a polymer called chromatin that also regulates gene expression. There are two major chromatin states: Euchromatin is open or accessible and thus, genes can be activated. In contrast, heterochromatin is closed or inaccessible and therfore, genes are repressed. Despite rapid advances in sequencing technologies, the repetitive content of the genome remained an enigma due to its redundancy, which interferes with standard protocols. However, understanding the chromatin organization and transcriptional regulation of such regions is important, as they contain many potentially harmful genetic elements called (retro)transposons. Transposons, or 'jumping genes' are mobile genetic elements that can multiply and insert at various positions in the DNA, which can lead to gene disruptions. Moreover, transcriptional de-regulation of retrotransposons is a hallmark of several human cancers.
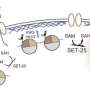
A research team led by Prof. Dr. Thomas Jenuwein, director at the MPI-IE, now investigated these genome areas in mouse stem cells using heterochromatin factors. They conducted genome-wide mapping for the enzyme 'histone methyltransferase Suv39h', one of the most prominent enzymes involved in heterochromatin formation, and its catalytic product, methylation of the packaging protein histone H3 at lysine 9. Subsequent refined bioinformatic analyses allowed the identification of repeat-associated heterochromatin. "Developing new bioinformatic tools for the analysis of repetitive regions was the key to this study," says Jenuwein. " This was only possible due to the close interaction of our wet lab scientists with the bioinformatics unit of Thomas Manke at the MPI-IE."
The new approach showed that repeat-associated heterochromatin has an important function to safeguard the genomic integrity of embryonic mouse stem cells. The Suv39h enzyme mainly targets retrotransposons of the LINE and LTR family. Surprisingly, Suv39h specifically silences only intact, and therefore potentially active, elements, which barely make up 2% of all retrotransposons. Upon loss of Suv39h, only these elements become transcriptionally upregulated. "Initially, we were surprised to see that such a small fraction of the repetitive genome responds to epigenetic regulation" explains co-first author Dr. Inti De La Rosa-Velazquez. "However, it makes sense to only regulate the potentially harmful elements. Now, we are very interested to dissect the molecular mechanism that allows the Suv39h enzymes to identify active retrotransposons."
In addition to stem cells, the researchers also investigated repetitive regions in more differentiated cells. In neural precursor cells or fibroblasts, retrotransposons lose histone methylation and instead accumulate DNA methylation, another epigenetic mechanism for gene silencing. This suggests that Suv39h-dependent repression is specific for stem cells. "Previous studies had shown that DNA methylation is the primary silencing mechanism for retrotransposons in differentiated cells", elaborates Jenuwein. "However, in stem cells which show reduced levels of DNA methylation, chromatin-based mechanisms were expected to safeguard retrotransposon repression. With our work on the Suv39h enzymes we have now identified such a pathway."
Provided by Max Planck Society