January 28, 2014 feature
Unpacking the past: Identifying a key evolutionary step in E. coli metabolism
.png)
(Phys.org) —Evolution is a process that takes place over long periods of time over which genetics and ecology may interact, producing novel phenotypic traits. Researchers previously found that after roughly 31,500 generations had passed in a laboratory evolution experiment a rare metabolic innovation emerged in which Escherichia coli developed the ability to metabolize citrate (Cit+) in its growth medium. Through genetic analysis with a recursive genomewide recombination and sequencing method (REGRES), scientists at University of Texas, Austin have now identified a key mutation that they say demonstrates how improvement of an emergent trait can be as important to its eventual success as earlier mutations or environmental conditions that may have been necessary for it to initially evolve.
Dr. Jeffrey E. Barrick discussed the paper that he, Graduate Research Assistant Erik M. Quandt and their co-authors published in Proceedings of the National Academy of Sciences. "REGRES is a combination of tried-and-true genetic methods, known for decades, including conjugation," Barrick tells Phys.org. (Conjugation is a form of bacterial pseudo-sex for exchanging genes or mutations, in which a portion of the chromosome of one bacterium is transferred into another.) "The new aspect in REGRES," Barrick continues, "was creating a way for this process to be used serially – that is, to keep performing this transfer into a 'blank' ancestral genome, starting from a genome that had many evolved genetic changes." This so-called backcrossing procedure sequentially removed all mutations that weren't required for surviving the growth conditions used, which required being able to efficiently utilize citrate as a nutrient. The other technology that enabled REGRES, Barrick notes, was next-generation DNA sequencing, which allowed the scientists to fully track which mutations were removed by not being transferred at each step.
In one of the study's key findings, the researchers identified a mutation that converts a rudimentary form of the innovation into a refined trait that confers a decisive competitive advantage, by dissecting the genetics of this trait using REGRES. "We were very surprised that after starting with a collection of more than 70 mutations in the evolved Cit+ strain – most of which were related to improving growth on glucose as a carbon source – we were able to find just two mutations that conferred the ability to utilize citrate." Barrick explains that this was unexpected due to very compelling evidence that mutations which occurred before citrate utilization arose in this population made certain E. coli in this population more likely to be able to evolve the ability to use citrate. This evidence is from studies that re-played evolution from many genetic starting points in what he describes as "very elegant" experiments by Zachary Blount and Richard Lenski at Michigan State University1,2. In addition, Barrick states that the REGRES method was key to sifting through the more than 70 evolved mutations that could have been necessary for utilizing citrate.
"Our results imply that these earlier 'potentiating' mutations weren't absolutely necessary for expressing the beneficial Cit+ trait, as we first thought. Instead," Barrick points out, "they seem to make it possible for evolution to follow a tenuous path. We hypothesize that these mutations may actually be deleterious to E. coli growth without the earlier potentiating mutations rewiring bacterial metabolism in some unknown way." This would mean that if these mutations occurred in certain strain backgrounds (like the ancestor) that they would rapidly go extinct from the population – but with the potentiating mutations present, it appears that the first mutation that affected the CitT transporter protein and gave a weak ability to use citrate was ever-so-slightly beneficial for growth and survival.
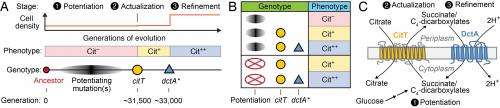
"Once this mutation was able to persist in the face of competition with other E. coli lineages in the population that were improving through other mutations," Barrick adds, "it gave enough time to get the second key mutation, which affects the DctA transporter protein. This mutation completed a cycle that enabled all of the citrate in the media to be efficiently utilized by this strain." This innovation gave the E. coli strain in question what Barrick describes as an "overwhelming" evolutionary advantage relative to its competitors that remained limited to competing for a lesser amount of the nutrient glucose.
An important conclusion of the study is that refinement of an emergent trait from a rudimentary form may be crucial to its evolutionary success. "Evolutionary innovations often seem to come out of nowhere, like the huge population expansion in this laboratory E. coli population when it evolved to be able to fully exploit citrate that had always been present in the growth medium for the preceding 15 years," Barrick tells Medical Xpress. "Our study shows that earlier mutations that may not have immediately obvious effects can be important for setting the stage for evolutionary breakthroughs. It's this exploratory mode of evolution – where there may not be any immediately obvious effects until a whole string of complementary mutations are put together – that can surprise us when we watch evolution happen in real time."
More specifically, the study's results imply that the DctA mutation was important for improving this rudimentary trait to enable full citrate utilization. "Once the REGRES results implied that only the DctA and CitT mutations were common to all backcrossed genomes, we were able to take the ancestral strain and add just those two mutations to it. Since this recapitulated a very strong ability to use citrate, we concluded that these were the key mutations required for the Cit+ phenotype." Moreover, he adds, the timing of these two mutations in the original population was also coincident with the earliest known weak Cit+ utilizers (CitT mutation) and a population expansion where E. coli with the refined Cit+ trait took over the population (DctA mutation) and grew to a much higher density than was possible without using this additional nutrient source.
Moving forward, Barrick says, the researchers remain very interested in identifying the potentiating mutations and/or ecological conditions that enabled the weakly Cit+ E. coli to persist in the population long enough for Cit+ refinement causing the population expansion to occur. "To solve this puzzle, we're trying to more precisely determine the order of the more than 70 mutations in the Cit+ strain that was examined in this study." They're doing this, he explains, by going back into the frozen "fossil record" of this population and sequencing the genomes of more bacteria. "We hope to eventually find a context of earlier mutations that makes the actualizing CitT mutation beneficial enough to explain why it did not go extinct when it occurred." Since the fitness effects of these mutations are much more subtle, Barrick adds, this is proving to be difficult.
"In the short term," he continues, "we're also following up on an interesting story related to a mutation that we discovered as a side effect of performing REGRESS. We found that a new mutation appeared during our backcrossing procedure that was not present in the Cit+ strain that we started with. It turns out that similar mutations eventually appeared many times in the actual long-term E. coli evolution experiment population, so it was beneficial for further refinement of the Cit+ trait to get one of these mutations." Interestingly, this mutation appears to reverse the effects of an earlier mutation that is present in these genomes, which appeared before citrate utilization ever arose. "We therefore have a case where becoming Cit+ changed E. coli metabolism so much that it made it beneficial to reverse the trajectory of earlier evolution. This sort of rewiring is a hallmark of evolutionary innovation."
Along with many other researchers Barrick and his co-authors continue to develop improved tools like REGRES to remix and edit the sequences of bacterial genomes. "For example," he illustrates, "it can still be difficult to go into the lab and reconstruct complex sequence rearrangements in a genome like the one that turned on expression of the CitT transporter."
Barrick notes that other areas of research might benefit from their study. "The REGRES methodology has several possible applications. It can be used, as in our study, to find which of many possible mutations in an evolved E. coli strain contribute to a complex phenotypic trait. One could also use the same approach to hybridize the genomes of two E. coli strains that have been evolved or engineered to have an industrially useful property, such as producing increased amounts of a biofuel. The hybrid genomes may acquire a collection of some mutations from each 'parent' that performs better than either of the original strains. A related use," he concludes, "would be to subtract deleterious mutations that sometimes accumulate over time in bacterial strains from a genome so that they're not a drag on its growth or production capacity."
More information: Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli, Proceeding of the National Academy of Sciences Published online before print on December 30, 2013, doi:10.1073/pnas.1314561111
Related:
1Genomic analysis of a key innovation in an experimental Escherichia coli population, Nature 489(7417):513–518 (2012), doi:10.1038/nature11514
2Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli, Proceeding of the National Academy of Sciences, 105(23):7899–7906 (2008), doi:10.1073/pnas.0803151105
Journal information: Proceedings of the National Academy of Sciences , Nature
© 2014 Phys.org. All rights reserved.