Sorting good germs from bad, in the bacterial world

(Phys.org) —Arizona State University scientists have developed a microfluidic chip, which can sort good germs from bad.
Your intestines are home to about 100 trillion bacteria. That's more than the number of cells that comprise the entire human body. Armies of bacteria sneak into our bodies the moment we are born, uninvited but necessary guests.
For the most part, these bacteria are industrious and friendly. Some of them are even beneficial, helping with digestion and producing vitamins. A few miscreants, though, will kill us if we let them stay.
Sometimes the difference between harmless and harmful is miniscule. Take E. coli for instance. Billions of E. coli organisms live in the average person's intestines. They go about their business causing no trouble whatsoever. However, one particular strain of E. coli, O157:H7, causes about 2,000 hospitalizations and 60 deaths in the U.S. every year. The differences between this strain and others are detectable only at the molecular level.
But how do we separate friend from foe? Determining whether or not bacteria are harmful usually requires growing cultures from food or infected patients. This is a time-consuming process that must be carried out in a laboratory. Since an estimated 9.4 million cases of food-borne illness occur each year in the U.S., we stand to gain much from new technologies that can rapidly identify microorganisms.
Scientists at Arizona State University's Department of Chemistry and Biochemistry, in the College of Liberal Arts and Sciences, have developed a new device that could significantly speed up the identification process for harmful bacteria and other microorganisms. The team, led by Professor Mark A. Hayes, hopes to create handheld, battery-operated devices that could deliver answers in minutes, instead of days.
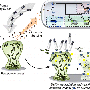
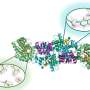
Identification takes place within a microscopically small channel in a chip made from glass or silicone polymer. The microchannel features saw-tooth shapes that allow researchers to sort and concentrate microbes based on their unique electrical properties.
The phenomenon that makes this work is called dielectrophoresis, which involves an applied voltage that exerts force upon the bacteria. This force acts like a coin-sorter, causing bacteria to become trapped at different points along the channel. Where they stop, and at what voltage, depends on their molecular and electrical properties.
Using this approach, Hayes's team including graduate student, Paul V. Jones, has separated extremely similar bacteria—pathogenic and nonpathogenic strains within the single species, E. coli. Their results have recently been published in "Online First" on SpringerLink and in the journal Analytical and Bioanalytical Chemistry.
"The fact that we can distinguish such similar bacteria has significant implications for doctors and health officials," says Hayes. He explains, "that scientists have struggled to find ways to rapidly identify bacteria. E. coli O157:H7 is very similar in size and shape to other subtypes of the bacteria. But unlike many of the others it has the ability to produce shiga-like toxin, a protein that breaks down blood vessel walls in the digestive tract."
Fortunately, all of these bacterial strains also possess subtle, but telltale differences in the proteins and other molecules that they express on their surface. According to Professor Hayes, dielectrophoresis is well suited to probe these phenotypic differences.
The researchers used an ordinary strain of E. coli along with two pathogenic varieties. They injected the cells into each channel and simply applied voltage to drive the cells downstream. The geometric features of the channel shape the electric field, creating regions of different intensity. This field creates the dielectrophoretic force that allows some cells to pass, while trapping others based on their phenotype.
So far, the device has only been used to test pure cultures of bacteria, but they hope soon to test complex mixtures of particles that are found in nature or the human body.
The next step is to create cheap, portable devices that would enable point-of-care or field based analysis. Such a device would require no time-consuming culturing or other tests, which would allow rapid response to disease or contamination, hopefully saving lives.
Journal information: Analytical and Bioanalytical Chemistry
Provided by Arizona State University