New database catalogs thousands of genetic variants in cassava—one of the world's primary food sources

Cassava is a woody shrub that is native to South America and is extensively cultivated in tropical regions worldwide, with an annual crop production of over 200 million tons. A team of researchers led by Tetsuya Sakurai from the RIKEN Center for Sustainable Resource Science has now completed the largest study to date of cassava DNA sequence variations.
The cassava plant, Manihot esculenta, has a tuberous root that serves as a primary food source for hundreds of millions of people, and the starch extracted from it is widely used in the food, paper and textile industries. Cassava is also highly resistant to drought, and its tubers can remain healthy in dry soil for up to several years. It therefore provides food security as it is a rich source of carbohydrate that can be drawn upon to prevent or relieve famine.
Sakurai and his colleagues retrieved more than 80,000 partial cassava DNA sequences from GenBank, a publicly available genetic sequence database run by the US National Institutes of Health. Using a computational approach to examine the sequences, they identified and characterized 10,546 DNA sequence variations called single nucleotide polymorphisms (SNPs), as well as 647 insertions and deletions.
They found that 62.7% of the SNPs occurred in protein-coding regions of the genome, and that genes conferring disease resistance contained a significantly higher ratio of 'non-synonymous' DNA sequence variations, which alter the coding sequence, than 'synonymous' variations, which do not.
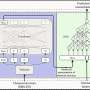
The researchers organized and integrated all of this information into the Cassava Online Archive, a comprehensive database that is freely available online at cassava.psc.riken.jp. The database can be searched by keyword, specific genetic variation, cassava variety and several other criteria. It also includes information about all the variants identified, such as their exact location on the genome and how they can be isolated.
Variations in protein-coding sequences of the genome probably have subtle influences on protein structure and function that are likely to play an important role in cassava's ability to adapt to environmental changes. The Cassava Online Archive could therefore help researchers to gain a better understanding of how the crop can survive in harsh conditions.
"The Cassava Online Archive is an ongoing project," says Sakurai. "We have plans to append gene expression profile data and aim to develop the database as a one-stop shop for cassava research."
More information: Sakurai, T., et al. Genome-wide discovery and information resource development of DNA polymorphisms in cassava, PLoS ONE 8 e74056 (2013). dx.doi.org/10.1371/journal.pone.0074056
Journal information: PLoS ONE
Provided by RIKEN









.jpg)