Researchers identify key cellular organelle involved in gene silencing
RNA molecules, made from DNA, are best known for their role in protein production. MicroRNAs (miRNAs), however, are short (~22) nucleotide RNA sequences found in plants and animals that do not encode proteins but act in gene regulation and, in the process, impact almost all biological processes—from development to physiology to stress response.
Present in almost in every cell, microRNAs are known to target tens to hundreds of genes each and to be able to repress, or "silence," their expression. What is less well understood is how exactly miRNAs repress target gene expression.
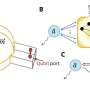
Now a team of scientists led by geneticists at the University of California, Riverside has conducted a study on plants (Arabidopsis) that shows that the site of action of the repression of target gene expression occurs on the endoplasmic reticulum (ER), a cellular organelle that is an interconnected network of membranes—essentially, flattened sacs and branching tubules—that extends like a flat balloon throughout the cytoplasm in plant and animal cells.
"Our study is the first to demonstrate that the ER is where miRNA-mediated translation repression occurs," said lead researcher Xuemei Chen, a professor of plant cell and molecular biology and a Howard Hughes Medical Institute-Gordon and Betty Moore Foundation Investigator. "To understand how microRNAs repress target gene expression, we first need to know where microRNAs act in the cell. Until now no one knew that membranes are essential for microRNA activity. Our work shows that an integral membrane protein, AMP1, is required for the miRNA-mediated target gene repression to be successful. As AMP1 has counterparts in animals, our findings in plants could have broader implications."
Study results appear today in the journal Cell.

Simply put, DNA makes RNA, and then RNA makes proteins. Specifically, RNA encodes genetic information that can be "translated" into the amino acid sequence of proteins. But noncoding RNAs—RNAs that do not encode proteins—are increasingly found to act in numerous biological processes. MicroRNAs are a class of noncoding RNAs whose main function is to downregulate gene expression.
Research on miRNAs has increased tremendously since they were first identified about 20 years ago. In the case of diseases, if some genes are up- or down-regulated, miRNAs can be used to change the expression of these genes to fight the diseases, thus showing therapeutic potential.
MicroRNAs are known to regulate target genes by two major modes of action: they either destabilize the target RNAs, leading to their degradation, or they do not impact the stability of the target RNAs, but simply prevent them from being translated into proteins—a process known as translation inhibition. The end result of translation inhibition is that the genes do not get expressed. Just how miRNAs cause translational inhibition of their target genes is not well understood.
"We were surprised that the ER is required for the translational inhibition activity of miRNAs," Chen said. "This new knowledge will expedite our understanding of the mechanism of gene silencing. Basically, now we know where to look: the ER. We also suspect it is the rough ER portions that are involved."
Chen explained that the ER has two types: rough and smooth. Rough ER, which synthesizes and packages proteins, looks bumpy; smooth ER, which acts in lipid synthesis and protein secretion, resembles tubes. The ER protein AMP1, she said, is anchored in the rough ER.
"My lab has been conducting research on AMP1 for many years," she said. "And it's this protein that drew our attention to the ER. First, we realized that AMP1 is involved in miRNA-mediated translational inhibition. Then, since we already knew that AMP1 is localized in the rough ER, we shifted our focus to this organelle."
Next, her lab will attempt to crack the mechanism of miRNA-mediated translational inhibition. They will investigate, too, how miRNAs are recruited to the ER.
Journal information: Cell
Provided by University of California - Riverside