Researchers discover a new biochemical pathway to producing ribose
A researcher from the University of Toronto Faculty Of Medicine’s Donnelly Centre for Cellular and Biomolecular Research is part of an international team which has discovered a new metabolic pathway for the production of ribose, a key component of RNA and DNA, providing new insight into the fundamental pathways that organisms utilize to grow and divide.
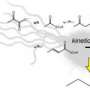
This new pathway, termed “riboneogenesis”, is the first new core metabolic pathway to be discovered in more than 30 years, and its discovery will necessitate the rewriting of biochemistry textbooks. It was discovered and characterized by lead authors Drs. Michelle Clasquin, Joshua Rabinowitz, Amy Caudy and her colleagues using a combination of genetic, biochemical, and structural techniques. Central to this finding is a previously unknown enzyme, a sedoheptulose biphosphate named SHB17, that plays a critical role in directly connecting the digestion of sugar to the production of ribose.
The findings are published in the June 10 issue of the journal Cell, which covers cutting-edge research in many areas of experimental biology. Caudy - an assistant professor at the Donnelly Centre -- and her colleagues used baker’s yeast as a model organism to identify and characterize a previously unknown connection between the breakdown of glucose and other sugars and the synthesis of ribose, a key component of DNA and RNA.
“With the availability of the complete genome sequences of organisms from yeast to man, we now know the full catalogue of genes for each. However, such lists only hint at how organisms process nutrients. Each cell in our bodies must take in nutrients to maintain itself. If so directed by environmental cues, our cells alter their metabolism in order to produce the building blocks necessary to divide,” said Caudy.
“I am working to characterize the chemical reactions carried out by enzymes of unknown function and place them in context of cellular metabolism and disease. I am working with my co-authors to more fully understand the role of riboneogenesis and to identify other new metabolic pathways, their metabolic intermediates and other signalling molecules.”
In the present work, Ribogenesis in Yeast, Clasquin et al, Cell 10 June 2011, Caudy and colleagues used computational predictions from genome-wide data to direct high-resolution mass spectrometric analysis of a previously unknown enzyme, YKR043c. These experiments were performed as part of an international multi-institution collaboration combining leading researchers at Princeton University (Rabinowitz Lab), the University of Tennessee (Campagna lab), and the University of Toronto (Yakunin lab). The Caudy and Rabinowitz groups provided genetic and biochemical evidence for this novel pathway, assisted by synthetic chemistry performed by the Campagna lab. The Yakunin lab caught the enzyme in the act of catalyzing the newly-described reaction using x-ray crystallography.
Two routes for ribose synthesis have been known for more than half a century, where the main route consumes oxygen in the synthesis of this critical compound. A third route, the riboneogenic pathway discovered in this study, enables cells to generate ribose in a fashion that does not affect cellular oxidization state. The discovery of this pathway has far-reaching implications for understanding how cells grown in low oxygen conditions are able to replicate their DNA and divide. This work is an important step to a more complete understanding of the biochemical processes within a variety of oxygen-starved cells, ranging from industrial fermentation to rapidly growing tumours.
“There are many chemical reactions that are known to occur as cells break down nutrients, but there also still new reactions to be discovered ” said co-lead author, Joshua Rabinowitz of the Lewis -Sigler Institute for Integrative Genomics and the Department of Chemistry at Princeton. “This collaboration with Dr. Amy Caudy, taking advantage of technological advances in mass spectrometry, is revealing a spectrum of reactions that will substantially alter the canonical charts of metabolism.”
Provided by University of Toronto