This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
The genetic interplay in Impatiens downy mildew: A transcriptome-based approach to enhancing disease resistance
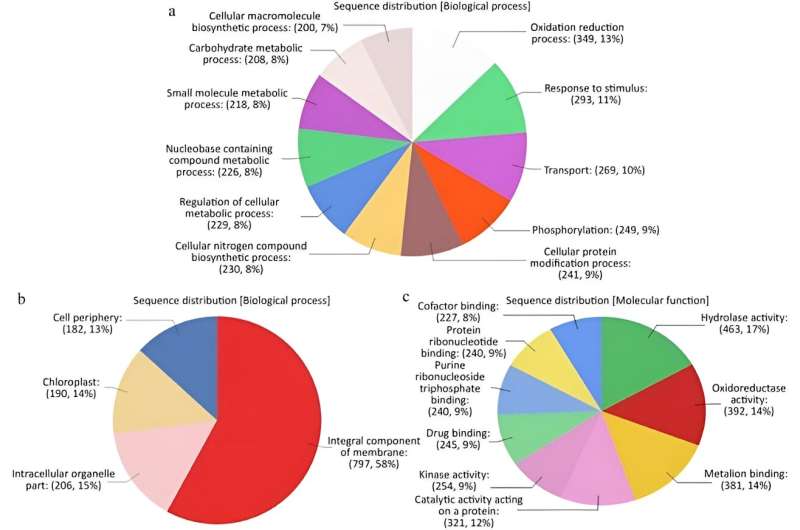
A research team has meticulously analyzed the transcriptional response of Impatiens walleriana to Plasmopara obducens infection, revealing between 3,000 and 4,500 differentially expressed transcripts at various stages of the disease.
Through this study, they have constructed a de novo transcriptome assembly that identified key genes involved in the plant susceptibility, including several stress response genes, receptor-like kinases (RLKs), and resistance genes. This investigation not only enhances our understanding of the molecular interactions between I. walleriana and its pathogen, but also provides a fundamental resource for future efforts to engineer disease resistance in this economically significant ornamental crop.
Downy mildews, including impatiens downy mildew (IDM), pose severe threats to both agronomic and ornamental crops, significantly impacting industries like floriculture. The pathogen, Plasmopara obducens, is a recent emergence and has had a major economic impact since it was first detected in cultivated Impatiens walleriana in the United States in 2004.
Current research has begun to unravel the genomic basis of the host and pathogen, yet gaps remain in understanding the complete host-pathogen interactions and developing IDM-resistant cultivars.
The study published in Ornamental Plant Research on 27 March 2024, dives into the transcriptome analysis of I. walleriana during its infection by P. obducens, aiming to pinpoint candidate genes linked to susceptibility. This research is pivotal in guiding future breeding efforts to enhance resistance against IDM, responding to strong consumer demand for robust I. walleriana cultivars.
More information: Stephanie Suarez et al, Transcriptional profiling of Impatiens walleriana genes through different stages of downy mildew infection reveals novel genes involved in disease susceptibility, Ornamental Plant Research (2024). DOI: 10.48130/opr-0024-0006
Provided by Chinese Academy of Sciences