This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Microalgae with unusual cell biology could lead to improved understanding of harmful algal blooms
What are the molecular processes in a unicellular marine algae species that can cause harmful algal blooms? A research team led by microbiologist Prof. Dr. Ralf Rabus from the University of Oldenburg (Germany) has conducted the first detailed analyses of the unusual cell biology of Prorocentrum cordatum, a globally widespread species of the dinoflagellates group, using both advanced microscopic and proteomics approaches.
As the team reports in the journal Plant Physiology, the photosynthesis process in these microorganisms is organized in an unusual configuration which may help them to better adapt to the changing light conditions in the oceans. The results of the study could lead to an improved understanding of the incidence of harmful algal blooms, which may be becoming more frequent due to climate change.
Dinoflagellates are important organisms in both marine and freshwater ecosystems. These unicellular organisms make up a substantial proportion of free-living phytoplankton, which forms the basis of the food web in oceans and lakes. Some species, including Prorocentrum cordatum, can proliferate in warm, nutrient-rich waters and form harmful algal blooms.
"We studied this organism because despite its environmental relevance, its cell biology and metabolic physiology are still poorly understood," said Rabus. In addition to studying photosynthesis in the microalgae, the researchers also examined the structure of their cell nuclei and their response to heat stress in collaboration with teams from the Universities of Hanover, Braunschweig, and Munich and set out the findings in two other recently published papers.
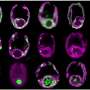
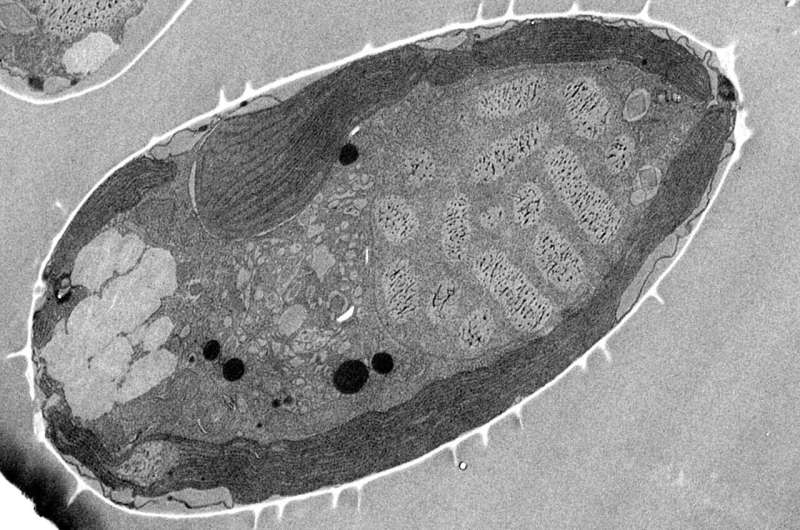
Using a powerful scanning electron microscope with a focused ion beam at the Ludwig-Maximilians-Universität Munich, the team headed by Rabus and lead author Jana Kalvelage from the Institute of Chemistry and Biology of the Marine Environment (ICBM) was able to reconstruct the three-dimensional architecture of the chloroplasts, where photosynthesis takes place.
The scientists were able to generate around 600 image layers of a single algae cell and then combine the sections to create a three-dimensional, high-resolution spatial image of the oval-shaped single-celled organisms, which are generally around 10 to 20 thousandths of a millimeter long. The analysis revealed that Prorocentrum cordatum have only a single barrel-like chloroplast that takes up 40 percent of their cell volume.
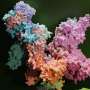
Proteomic (protein) analyses then revealed marked differences between the photosynthetic apparatus of the microalgae and that of Arabidopsis thaliana, a well-studied model plant in genetics research. In both species, photosynthesis takes place in complex protein structures embedded in the chloroplast's extensive membrane system.
However, in Prorocentrum cordatum the team observed that the conversion of solar energy into biochemical energy takes place in a single large structure consisting of numerous proteins, known as a "megacomplex," whereas in the chloroplasts of the plant species the different steps of photosynthesis occur in spatially separated structures.
The team also reported that P. cordatum uses a large number of different pigment-binding proteins to capture solar energy efficiently. "This diversity is a special adaptation to the changing light conditions to which the organism is exposed in the oceans," Rabus explained.
Two other studies published last year highlight the microalgae's unusual biology: in the first, a German-Australian team of which the ICBM researchers were also members found that the organisms have a very large genome with twice as many base pairs as in humans. The team also discovered that the algae change their metabolism, and their rate of growth decelerates in response to heat stress.
In a second paper published in mSphere, the team led by Rabus and Kalvelage described the cell nucleus in greater detail, reporting that P. cordatum has 62 chromosomes, an unusually high number that fills almost the entire cell nucleus. The function of a large proportion of the nuclear proteins that were identified by the researchers is currently unknown, the team observed.
"We have investigated how this important microalgae functions at the molecular level. These findings form the basis for a better understanding of its role in the environment," Rabus stressed. Further investigations could provide answers to questions such as how the organism's metabolism reacts to other stress factors—and why the species is able to adapt to such a wide range of environmental conditions, from those in the tropics to those in temperate climates, he explained.
More information: Jana Kalvelage et al, Conspicuous chloroplast with light harvesting-photosystem I/II megacomplex in marine Prorocentrum cordatum, Plant Physiology (2024). DOI: 10.1093/plphys/kiae052
Jana Kalvelage et al, The enigmatic nucleus of the marine dinoflagellate Prorocentrum cordatum, mSphere (2023). DOI: 10.1128/msphere.00038-23
Provided by Carl von Ossietzky-Universität Oldenburg