This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Super-resolution microscopy harnesses digital display technology
In the ever-evolving realm of microscopy, recent years have witnessed remarkable strides in both hardware and algorithms, propelling our ability to explore the infinitesimal wonders of life. However, the journey towards three-dimensional structured illumination microscopy (3DSIM) has been hampered by challenges arising from the speed and intricacy of polarization modulation.
Enter the high-speed modulation 3DSIM system "DMD-3DSIM," combining digital display with super-resolution imaging, allowing scientists to see cellular structures in unprecedented detail.
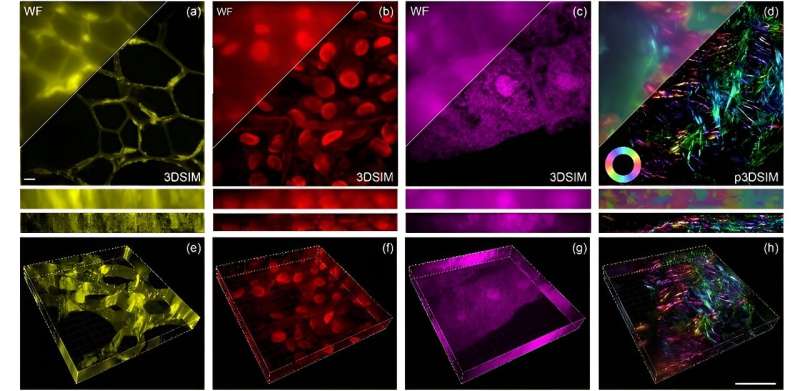
As reported in Advanced Photonics Nexus, Professor Peng Xi's team at Peking University developed this innovative setup around a digital micromirror device (DMD) and an electro-optic modulator (EOM). It tackles resolution challenges by significantly improving both lateral (side-to-side) and axial (top-to-bottom) resolution, for a 3D spatial resolution reportedly twice that achieved by traditional wide-field imaging techniques.
In practical terms, this means DMD-3DSIM can capture intricate details of subcellular structures, such as the nuclear pore complex, microtubules, actin filaments, and mitochondria in animal cells. The system's application was extended to study highly scattering plant cell ultrastructures, such as cell walls in oleander leaves and hollow structures in black algal leaves. Even in a mouse kidney slice, the system revealed a pronounced polarization effect in actin filaments.
An open gateway to discovery
What makes DMD-3DSIM even more exciting is a commitment to open science. Xi's team has made all the hardware components and control mechanisms openly available on GitHub, fostering collaboration and encouraging the scientific community to build upon this technology.
The DMD-3DSIM technique not only facilitates significant biological discoveries but also lays the groundwork for the next generation of 3DSIM. In applications involving live cell imaging, advancements in brighter and more photostable dyes, denoising algorithms, and deep learning models based on neural networks promise to enhance imaging duration, information retrieval, and real-time restoration of 3DSIM images from noisy data. By combining hardware and software openness, the researchers hope to pave the way for the future of multidimensional imaging.
More information: Yaning Li et al, High-speed autopolarization synchronization modulation three-dimensional structured illumination microscopy, Advanced Photonics Nexus (2023). DOI: 10.1117/1.APN.3.1.016001
Provided by SPIE