This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
The effects of primer pairs, PCR conditions, and peptide nucleic acid clamps on plant root fungal diversity assessment
Fungi are frequently found both around and within plant tissues (especially in roots) and are involved in both plant nutrient acquisition and resistance to pathogens. Thus, characterizing the diversity and composition of plant-associated fungal communities has been a growing interest in recent years.
High-throughput sequencing (HTS), also called metabarcoding, has become a prominent tool to assess complex microbial communities from environmental samples. However, HTS applied to plant-associated fungal communities is challenging due to plant and fungal DNA co-amplification. Fungal-specific primers, Peptide Nucleic Acid (PNA) clamps, or adjusting PCR conditions are described as efficient approaches to limit plant DNA contamination.
This study, led by Dr. Coralie Bertheau (Université de Franche-Comté), evaluated the combined effects of primer pairs, associated annealing temperature (Ta), and PNA clamps in determining the fungal community diversity and composition associated with plant roots.
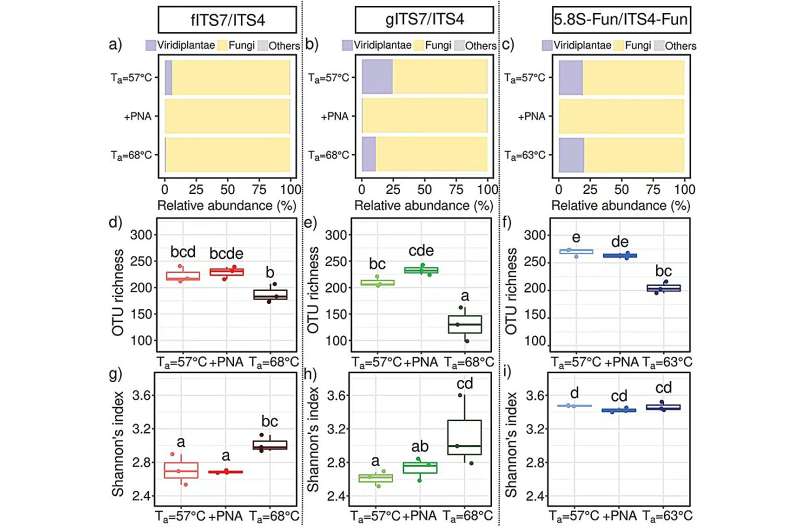
Three primers (fITS7/ITS4, gITS7/ITS4, and 5.8S-Fun/ITS4-Fun) targeting the ribosomal internal transcribed spacer (ITS) 2 region were evaluated alone or in combination with PNA clamps both on nettle (Urtica dioica) root DNA and a mock fungal community.
The team found that PNA clamps did not improve the richness or diversity of the fungal communities but increased the number of fungal reads. Among the tested factors, the most significant was the primer pair. Specifically, the 5.8S-Fun/ITS4-Fun pair exhibited a higher OTU richness but fewer fungal reads.
"The results demonstrated that the choice of primers is critical for limiting plant and fungal DNA co-amplification. PNA clamps increase the number of fungal reads when ITS2 is targeted but do not result in higher fungal diversity recovery at high sequencing depth. At lower read depths, PNA clamps might enhance microbial diversity quantification for primer pairs lacking fungal specificity," Dr. Coralie Bertheau said.
The work is published in the journal Mycology.
More information: Chloé Viotti et al, Primer pairs, PCR conditions, and peptide nucleic acid clamps affect fungal diversity assessment from plant root tissues, Mycology (2024). DOI: 10.1080/21501203.2023.2301003
Provided by Tsinghua University Press





















