July 7, 2023 feature
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Developing a human malaria-on-a-chip disease model
In a new report published on Scientific Reports, Michael J. Rupar, and a research team at Hesperos Inc., Florida, U.S., developed a functional, multi-organ, serum-free system to culture P. falciparum—a protozoan that predominantly causes severe and fatal malaria, in order to establish innovative platforms to develop therapeutic drugs.
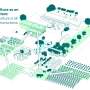
The platform contained four human organ constructs, including hepatocytes, splenocytes, endothelial cells, and recirculating blood cells, for interactions with the parasitic organism to simulate an infection. The team used two strains of P. falciparum; the 3D7 strain sensitive to chloroquine; a well-established anti-malarial drug, and the W2 strain resistant chloroquine. They maintained functional cells in healthy and diseased conditions for 7 days in the recirculating microfluidic model.
The scientists demonstrated an effective platform for therapeutic development where chloroquine treatment significantly decreased parasitemia in the 3D7 strain-constituent model. They used this setup for therapeutic index determination to evaluate off-target toxicity for anti-malarial treatment in a dose dependent manner. The outcomes can establish a new approach to evaluate anti-malarial therapies in a realistic human model that maintained blood circulation for 7 days.
The life cycle of Plasmodium falciparum
The parasitic lifecycle occurs in two stages: during a blood meal, sporozoites are released from the saliva of the mosquito, which then travel through peripheral blood circulation to the liver to replicate within hepatocytes. This results in an abundance of merozoites within a few days that are then released from the ruptured hepatocytes to navigate from the liver to the blood stream.
The lifecycle of the merozoites are asexual and erythrocytic, where the parasite develops by infecting blood cells. Rapid asexual replication fueled by the host's hemoglobin, caused merozoites to enter a ring stage, in 48 hours to then mature into trophozoites and schizonts, which continue to grow and replicate until they burst, releasing more merozoites to repeat the cycle of infection.
The development of anti-malaria strategies
Malaria is an epidemic that is on the rise from 2014 to 2020, with a steady increase in reports to emphasize the emergence of resistant strains across the years. As a result, bioengineers and life scientists are keen to develop new approaches for anti-malarial development to combat disease progression.
The Plasmodium parasite causes malaria and is transmitted by the female Anopheles mosquito. Of the variants, the falciparum species is the deadliest and is primarily responsible for a variety of severe malaria cases.
The World Health Organization aims to globally reduce malaria case incidences and mortalities by 90 percent by the year 2030. Researchers are keen to achieve this goal by investigating new platforms to study remedies for the disease. One such attempt is Rupar and colleague's development of a multi-organ, pre-clinical anti-malarial drug discovery platform that can establish and maintain healthy and disease conditions, as a cost-effective method to animal models.
The experimental setup
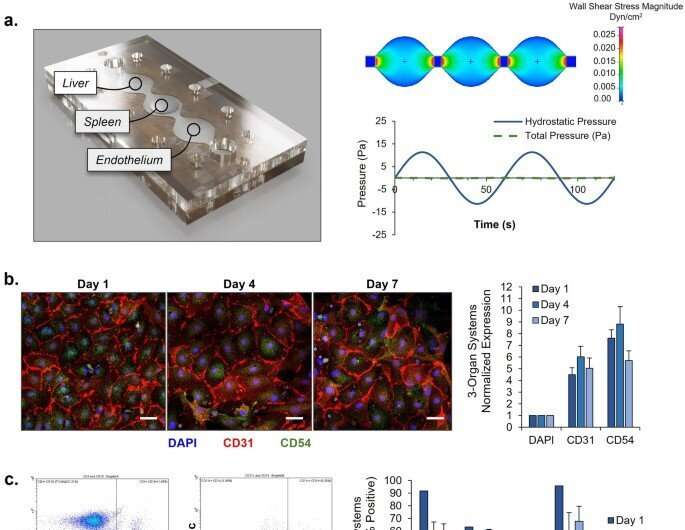
Rupar et al. used two strains of P. falciparum; a chloroquine sensitive and chloroquine resistant strain, which were successfully maintained across an 8-day period in the multi-organ model. The scientists established this instrument to assess malaria therapy in humans, to examine the efficiency and safety of the platform, while also exploring the safety of the therapeutic index for additional studies. Rupar and colleagues developed a microfluidic device containing three components to plate the liver, spleen, and endothelial cells.
The pumpless system introduced gravity-driven flow to the multi-organ system with sinusoidal rocking, with relevant physiological parameters. By engineering the malaria-on-a-chip model with microfluidics without pumps, they ensured recirculation of primary human red blood cells to simulate the preliminary stages of systemic infection with the P. falciparum parasite.
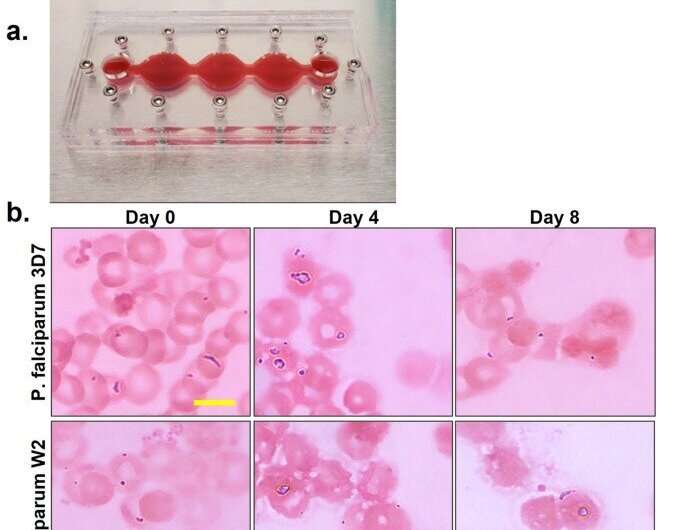
Malaria-on-a-chip
The scientists characterized the cells in the multi-organ system by assembling cells on chips without the parasite on the malaria-on-a-chip platform. They conducted the study for 7 days to determine cell viability, function, and appearance in a healthy microphysiological system across 7 days. The team then studied the lifecycle of the parasite within the instrument, and included two strains; the 3D7 strain and the W2 strain that are chloroquine sensitive and chloroquine resistant, respectively.
The researchers observed all stages throughout 8 days, to determine the level of parasitemia in each system.
Practicality of the organ-chip instrument
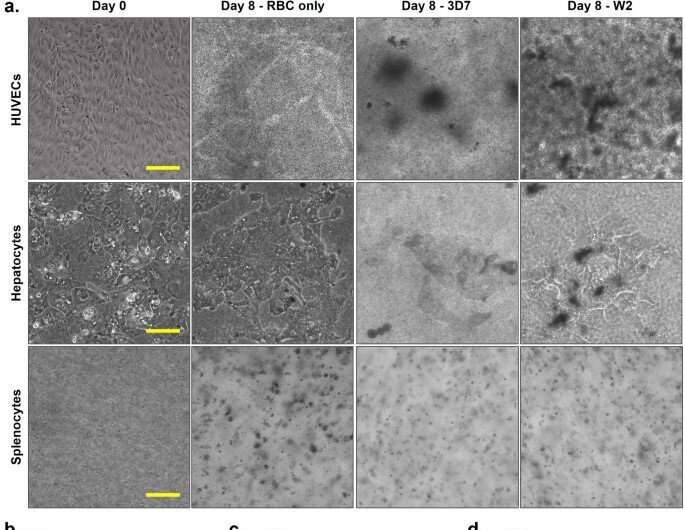
The team next studied the viability of functional cells in the organ-chip instrument where they contained red blood cells infected with either strain or uninfected blood for 8 days of culture. Using phase imaging, they captured the functional cells prior to assembly, and immediately after disassembly at day 8. Morphological changes indicated the disruption of cell viability upon infection with P. falciparum.
Rupar and colleagues monitored parasitemia every 12 hours and noted significantly lower parasite levels in the treatment group. Although this decrease in parasitic levels fluctuated between the drug sensitive and drug resistant system, ultimately the levels were significantly less in the chloroquine sensitive treatment group, when compared to the resistant systems. After disassembling the instruments, they continued to study the viability of the constituent cells.
Outlook
In this way, Michael J. Rupar and colleagues developed a four-organ malaria disease model representing a P. falciparum infection. Using the model, the team determined any off-target effects of chloroquine in the functionality or viability of organ constructs, and determined a therapeutic index for antimalarial therapy. The serum-free multi-organ construct contained a liver, spleen, and endothelium organs to present a cost-efficient approach to study antimalarial therapy.
This construct allows biochemists and bioengineers to study parasitic interactions in real-time within a microphysiological organ-chip environment, and identify foreseeable off-target effects of the therapeutic compounds.
More information: Michael J. Rupar et al, Development of a human malaria-on-a-chip disease model for drug efficacy and off-target toxicity evaluation, Scientific Reports (2023). DOI: 10.1038/s41598-023-35694-4
Louis H. Miller et al, The pathogenic basis of malaria, Nature (2002). DOI: 10.1038/415673a
Journal information: Scientific Reports , Nature
© 2023 Science X Network