This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New imaging technique can capture entire plant tissues in 3D
The cellular life inside a plant is as vibrant as the blossom. In each plant tissue—from root tip to leaf tip—there are hundreds of cell types that relay information about functional needs and environmental changes. Now, a new technology developed by Salk scientists can capture this internal plant world at an unprecedented resolution, opening the door for understanding how plants respond to a changing climate and leading to more resilient crops.
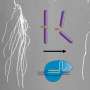
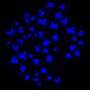
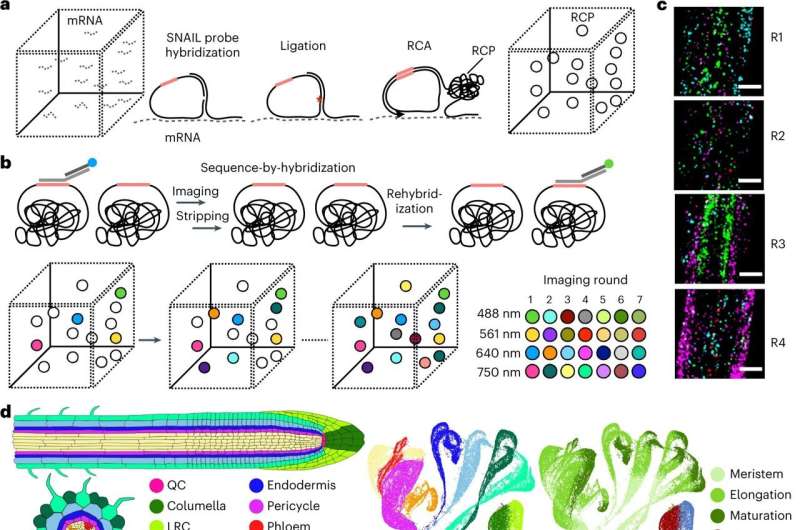
The method, called PHYTOMap, can capture entire plant tissues (like the whole root tip), instead of a small slice and provides insight into the complex biological conversations between cells that is difficult in two dimensions.
The method was detailed in Nature Plants on June 12, 2023, and the researchers expect PHYTOMap to be quickly popularized by the global scientific community.
"PHYTOMap allows us to examine dozens of plant genes and see which cells express those genes, how cells influence each other, and how tissue architecture influences those cells," says Salk Professor Joseph Ecker, director of the Genomic Analysis Laboratory and Howard Hughes Medical Institute investigator. "We can then use those answers to improve crops, predict plant reactions to climate change, and more."
Existing imaging techniques can only view a small number of genes in one type of plant tissue and requires altering the plants' genetic makeup (creating transgenic lines). PHYTOMap (short for plant hybridization-based targeted observation of gene expression map) allows researchers to study dozens of genes simultaneously without any time-consuming genetic manipulation of the plant.
"PHYTOMap was able to map various cell-type-specific genes in expected locations of root tips in 3D," says Tatsuya Nobori, a postdoctoral researcher in Ecker's lab. "Now, we can use PHYTOMap to ask more complex questions, like how do different cell types respond and react to each other and their environment?"
In addition to being powerful, PHYTOMap is also accessible—the technique used is relatively standard and the associated cost is relatively minimal.
"With PHYTOMap, we will be able to ask so many new biological questions. I can't wait to use the method to see how plants interact with surrounding microorganisms," says Nobori.
"PHYTOMap makes visualizing cells in plant tissues so much easier—no need to alter the plant's genetic makeup, no need to flag cells with colorful markers," says Ecker, who is also the Salk International Council Chair in Genetics. "I'm excited to see how PHYTOMap propels efforts to understand plant gene regulation during normal development and under various environmental conditions as well as how it may inform the optimization of agriculture."
In the future, the Ecker lab will use PHYTOMap to better understand the regulation of cell populations in various plant tissues to eventually engineer crops that are more resilient to climate change.
More information: Tatsuya Nobori et al, Multiplexed single-cell 3D spatial gene expression analysis in plant tissue using PHYTOMap, Nature Plants (2023). DOI: 10.1038/s41477-023-01439-4
Journal information: Nature Plants
Provided by Salk Institute