This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
AI method predicts how cells are organized in disease microenvironments
Cells in the human body, the building blocks of life, are arranged in a precise way. That's necessary because pathways and spaces provide a means for cells to communicate, collaborate and function within the specific tissue or organ. Changes in cell arrangement can lead to uncontrolled cell growth, cell death and diseases, including cancer.
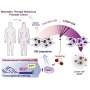
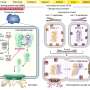
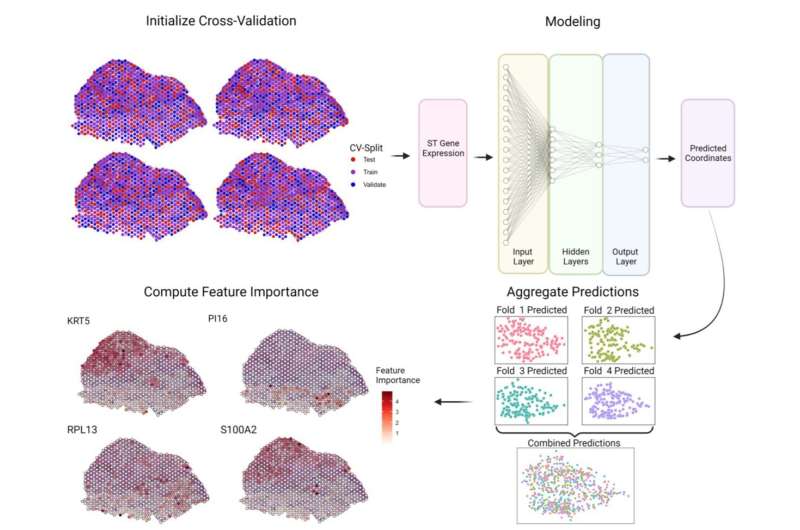
Scientists at the Mayo Clinic Center for Individualized Medicine and Mayo Clinic Comprehensive Cancer Center have developed an artificial intelligence method, called Spatially Informed Artificial Intelligence (SPIN-AI). This new deep-learning technique can analyze the genetic information of individual cells to reconstruct the precise layout of the cells in a tissue, without preexisting knowledge of how the cells are organized. The new study detailing SPIN-AI is published in Biomolecules.
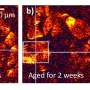
For the study, the team tested their novel SPIN-AI method on special data from a type of skin cancer called squamous cell carcinoma. This special data, known as spatial transcriptomic data, contains information on how thousands of genes are actively expressed within cells in their specific locations. By using this data, the team was able to create a detailed map revealing how the cells were organized and behaving in the diseased tissue.
"Knowing how cells are organized and communicate in diseases is paramount for a better understanding of what caused the disease, and for the formulation of new therapeutic designs," says Hu Li, Ph.D., a systems biologist and professor at Mayo Clinic's Department of Molecular Pharmacology and Experimental Therapeutics and Center for Individualized Medicine. Dr. Li is a co-lead author of the study.
The scientists say their findings could potentially pave the way for individualized treatments that target the specific cellular traits of each person.
"We selected this study because these data are widely available and include high-quality spatial transcriptomic data from many patients, which helps to benchmark our findings," says Cristina Correia, Ph.D., a pharmacology and oncology researcher in Mayo Clinic's Department of Molecular Pharmacology and Experimental Therapeutics and a co-lead author of the study. "We can now utilize these findings to further investigate how to help manipulate the cellular microenvironment to sustain and enhance drug responses."
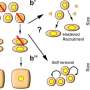
The researchers emphasize that their study was motivated by their hypothesis that specific genes exist that help predict the layout of cells.
Using their SPIN-AI innovation, the researchers discovered a new class of genes, which they have named "spatially predictive genes." These genes provide clues about where they are active or "turned on" within a cellular niche. By studying these genes, the researchers say they can gain insights into how cells are meticulously organized, and the role they are playing in the functioning of specific areas.
"Many people use AI simply for recognition tasks or classification tasks. But in our work, we show that we can make use of AI to make a new discovery that is driven by a hypothesis," says Choong Ung, Ph.D., a researcher in the Department of Molecular Pharmacology and Experimental Therapeutics. Dr. Ung is also a co-lead author of the study. "Asking the right questions and strategically crafting a well-designed training-testing procedure is crucial to harness the full power of AI, especially deep learning," Dr. Ung adds.
The team plans to continue working on ways to improve SPIN-AI to explore its capability to identify and distinguish different cell populations and investigate strategies to match them to therapeutic targets.
More information: Kevin Meng-Lin et al, SPIN-AI: A Deep Learning Model That Identifies Spatially Predictive Genes, Biomolecules (2023). DOI: 10.3390/biom13060895
Provided by Mayo Clinic