This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Label-free droplet-based methods improve rapid screening and sorting of bacteria
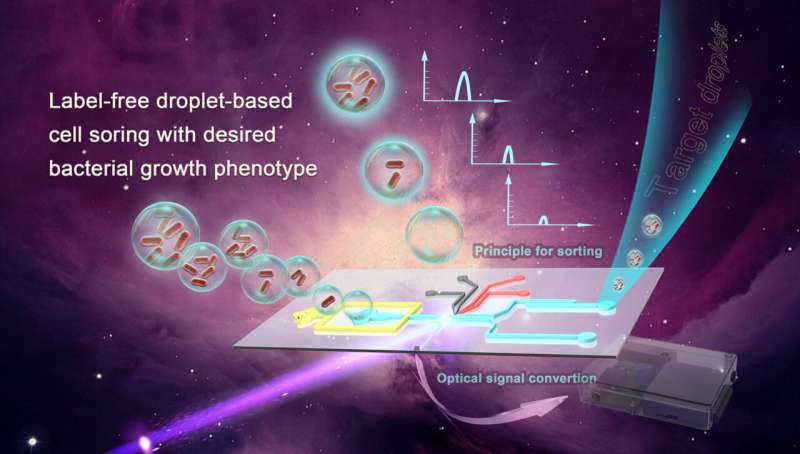
Effective, accurate and quick ways to screen and sort microbes are in short supply. Most methods available now rely on additional labeling steps to sort bacteria, which are typically time-consuming and cannot work well for industrial-scale breeding.
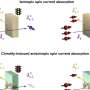
The need for accuracy and speed are met using a label-free droplet-based integrated microfluidic platform that screens bacterial growth based on phenotype, or observable characteristics. Better and quicker screening of bacterial growth can have considerable effects on the medical, pharmaceutical and agricultural industries.
Screening of bacteria by their observable characteristics may not seem possible, but the researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) determined how to best do this using a microfluidic system and the autofluorescent properties of bacteria.
The results were published in Sensors and Actuators B: Chemical on March 22.
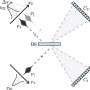
The main functions of this microfluidic platform are organized in a box, which houses the "detection area" the droplets will go through, along with photomultiplier tubes (PMT) and an optical fiber inserted through the chip. A light signal is transmitted to the PMT, and is sorted based on the intensity. There are parameters that can be set both high and low to sort out the target droplets easier.
"A flexible, label-free droplet-based detector allowing bacterial growth phenotype screening may help to expand the scope of rapid bacteria screening in various applications, especially in high-quality industrial breeding," said Ge Anle, researcher and first author of the study.
The researchers determined by using PMT that high densities of bacteria such as E. coli were able to be divided into a sorting channel when exposed to an 800V wave pulse for 5 to 20 milliseconds. Any droplets that did not make it to sorting thresholds were left to fall into the waste channel, and these were typically empty, or negative, droplets. Positive droplets, or droplets with bacteria in them, got deflected into a sorting channel since they were within the given threshold.
Results from the study indicated sorting efficiency of 95.3% when it comes to cell-containing (positive) droplets, and 91.7% of empty (negative) droplets successfully made it to the waste channel.
"We anticipate that the mini integrated microfluidic system will serve as a useful platform for further subsequent analyses, including antibiotic resistance, metabolic analysis and industrial strains growth phenotype screening," said Diao Zhidian, researcher and co-author of the study.
The microfluidic platform has many advantages compared to the way bacterial growth phenotype screening is done currently. Using an optical fiber to transmit light eliminates the need for a complex lighting system, and most of the core functions are fully integrated into the box structure.
"The use of determining phenotype using light properties but not relying on fluorescent labels is important, especially because many different cell types may not be compatible with sorting through the use of fluorescent labeling," said corresponding author Prof. Ma Bo, from Single-Cell Center of QIBEBT. "The last big advantage is the droplet method: isolating individual bacteria in a droplet can allow for better chances of proliferation, which is of utmost importance in fields involving large-scale industrial breeding of bacteria."
"We have developed Raman-based flow cytometry tools, such as FlowRACS which improves accuracy, throughput, and stability in profiling dynamic metabolic features of cells, to screen 'high-yield' strains rapidly without the need to label the cell with fluorescence probes. This newly developed microfluidic platform can further screen the 'fast-growing' strains rapidly, so as to achieve the goal of industrial microbial breeding," said Prof. Xu Jian, the head of Single-Cell Center of QIBEBT. "Next, we will further develop the key technologies and equipment platform for single-cell breeding to support the development of industrial biotechnology and synthetic biology."
More information: Anle Ge et al, Label-free droplet-based bacterial growth phenotype screening by a mini integrated microfluidic platform, Sensors and Actuators B: Chemical (2023). DOI: 10.1016/j.snb.2023.133691
Provided by Chinese Academy of Sciences