Study sheds light on phylogenomics and tree of life of flowering plants
Flowering plants (angiosperms) are the largest, most diversified, and most successful major lineage of green plants, with ~330,000 known species. In the past decade, the accelerated development of high-throughput sequencing technology has provided a great impetus for phylogenetic studies of angiosperms, and a large number of phylogenetic studies adopting hundreds to thousands of genes across a wealth of clades have emerged and ushered plant phylogenetics and evolution into a new era—the era of phylogenomics.
Prof. Li Dezhu's research team from the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS) has been engaged in angiosperm phylogenetic and evolutionary genomics for decades. Based on their own expertise and extensive literature survey, the researchers reviewed state of the reconstruction of the flowering plant tree of life, and discussed the major challenges faced by phylogenomic studies.
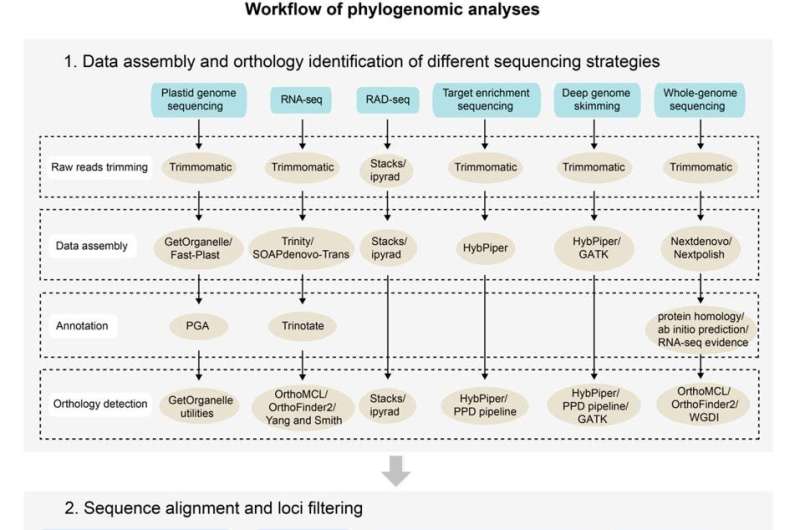
The researchers outlined the current sequencing strategies, including genome skimming, transcriptome sequencing, restriction site-associated DNA sequencing, target enrichment sequencing, deep genome skimming, and whole-genome sequencing, and discussed the advantages, shortcomings and potential applications of each. A workflow for plant phylogenomic analyses was provided for practical applications.
They also summarized recent developments in the use of genomic data for phylogenetic reconstructions of flowering plants at various taxonomic levels, and highlights their potential impact on taxonomic treatments.
They further discussed the major challenges, such as the adverse impact on orthology inference and phylogenetic reconstruction raised from systematic errors, whole-genome duplication, hybridization/introgression, and incomplete lineage sorting and made practical recommendations for phylogenomic studies of complex evolutionary lineages.
According to the researchers, as more and more unresolved relationships emerged across major flowering plant lineages, dichotomous branching phylogenetic trees, in some cases, may not be the best evolutionary model for the angiosperm tree of life, and it is hoped that development of network-based approaches would improve our ability to reconstruct phylogenetic relationships and infer evolutionary processes.
In addition, whole-genome sequencing is an inevitable trend in years to come, and the increasing accessibility of genomic data would continue to deepen our understanding of angiosperm evolutionary history;
The majority of phylogenomic studies focus on restricted geographic and taxonomic scopes, and a more balanced development through global collaboration is urgently needed.
Furthermore, integrated analysis of omics data coupled with morphological, ecological, and ultimately multi-disciplinary evidence is critical for a more complete understanding of plant evolutionary history.
"Phylogenomics and the flowering plant tree of life," has been published in the Journal of Integrative Plant Biology.
More information: Cen Guo et al, Phylogenomics and the flowering plant tree of life, Journal of Integrative Plant Biology (2022). DOI: 10.1111/jipb.13415
Provided by Chinese Academy of Sciences