Study reveals sequence and structural divergence in strawberry mitogenomes
Fragaria, known as strawberries, are among the most popular berry fruits and also the youngest domesticated plants. Southwest China is rich in wild strawberry germplasm resources, and has been recognized as an important distribution center of wild strawberry resources in the world.
The genus Fragaria has extensive interspecific hybridization and polyploidization, which makes the studies on the origin of organelle genomes and the evolution of nuclear-cytoplasmic interactions still largely unrevealed. This also hinders the effective evaluation and use of wild strawberry germplasm resources.
Compared with chloroplast genomes, plant mitochondrial genomes are generally characterized by complex and diverse structures, low substitution rate, frequent recombination, etc. It is considered as an important genetic system for studying the evolution of genome structure and function.
Previous studies have demonstrated that plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. Moreover, it has also been suggested that there may be different selection pressure or mutation repair mechanisms between the coding and noncoding regions of the mitochondrial genome, resulting in differences in the evolutionary rates.
To better understand the evolutionary features and mechanisms of plant mitochondrial genomes, researchers from the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS) have assembled 13 complete mitochondrial genomes from Fragaria wild species and studied the sequence variation characteristics (substitution rate, indels, inversions) of their coding and non-coding regions.
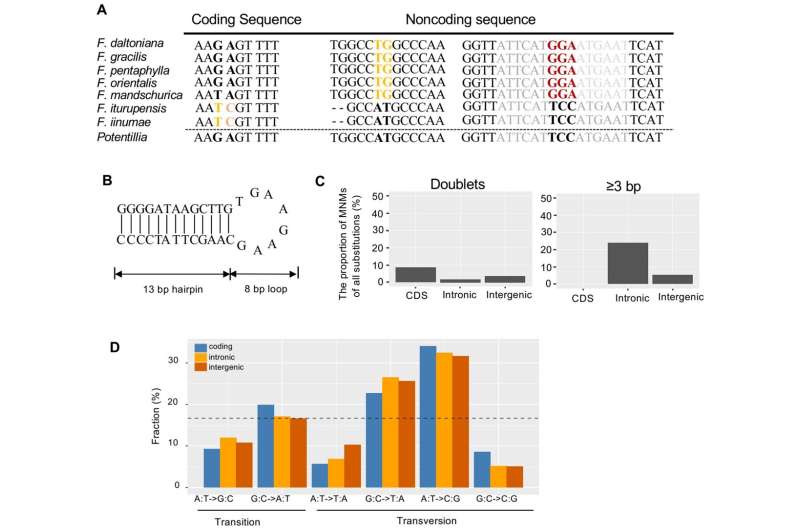
The researchers found that the genome-wide nucleotide substitution rates were generally consistent between noncoding regions and synonymous sites in Fragaria mitogenomes, but indels and inversions occurred more frequently in non-coding regions.
In addition, Fragaria mitochondrial genomes experienced frequent rearrangements, so the genomic structure varies obviously even among closely related species.
This study indicates that there are a large number of microinversion-mediated multinucleotide substitutions in Fragaria mitochondrial genomes, and these continuous mutation sites adversely affect local mutation rates and phylogenetic signal. Moreover, a gain-and-loss model can explain the general lack of homology among plant mitogenomes.
Results have been published in New Phytologist.
More information: Weishu Fan et al, Fragaria mitogenomes evolve rapidly in structure but slowly in sequence and incur frequent multinucleotide mutations mediated by micro‐inversions, New Phytologist (2022). DOI: 10.1111/nph.18334
Journal information: New Phytologist
Provided by Chinese Academy of Sciences





















