How does an intestinal microbe become a pathogen?
The bacterium Escherichia coli is found in the human intestine, and elsewhere. There it is harmless, but in certain conditions it can become a pathogen. It can cause bladder infections or even sepsis. A team of researchers led by RESIST professor Marco Galardini at TWINCORE, together with colleagues from the medical faculty of the University of Paris, has now investigated whether certain genes of the bacterium are associated with the severity of the diseases caused. They have now published their findings in the journal PLOS Genetics.
Escherichia coli, often abbreviated as E. coli, is part of the human intestinal flora. As a so-called commensal, it does not normally cause any damage there. However, it can become a pathogen in the gut and in other organs. In the urogenital tract, for example, E. coli causes bladder infections, and in the bloodstream it can cause sepsis. Blood poisoning is a dreaded consequence of bacterial infections and can even be fatal in 10 to 30 percent of cases. Until now, it was not possible to predict how severe such an infection would be based on the genetic makeup of the pathogen.
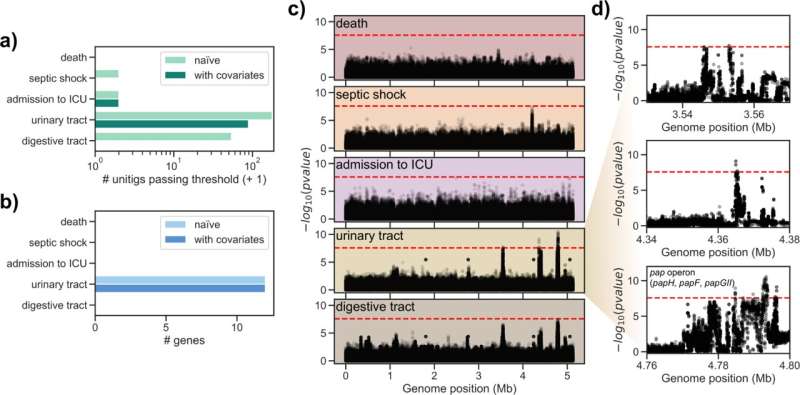
Researchers from TWINCORE in Hannover have now analyzed whether certain E. coli genetic variants are associated with a more severe course. "We carried out a so-called genome-wide association study," says Marco Galardini, head of the RESIST research group Systems Biology of Microbial Communities. "For this, we sequenced bacterial samples from two large patient studies and correlated them with the course of the infection." Characteristics such as age, gender or known previous diseases were also included in the analysis.
Galardini's team was unable to identify genes that determine the severity of the disease. However, they made another interesting discovery: "A certain gene cassette was clearly associated with infections that started in the urinary tract," says Galardini. From this, a strategy for avoiding life-threatening diseases can be derived. "In future, one could sequence the pathogens in the case of a bladder infection and then decide whether the drug treatment should be adjusted as a precaution," says Galardini.
The fact that the researchers could not prove a connection between the genome of the bacteria and the course of the disease does not necessarily mean that there is none. "It could just as well be that the number of samples we studied was too small," says Galardini. "A simulation showed that ten times the number of samples would be needed to detect or exclude the link with more confidence."
That is why Galardini and his French cooperation partners are preparing a more extensive follow-up study. They have already submitted an application for the necessary funding.
More information: Erick Denamur et al, Genome wide association study of Escherichia coli bloodstream infection isolates identifies genetic determinants for the portal of entry but not fatal outcome, PLOS Genetics (2022). DOI: 10.1371/journal.pgen.1010112
Journal information: PLoS Genetics
Provided by Zentrum für Experimentelle und Klinische Infektionsforschung