The molecular evolution of the aconitase superfamily
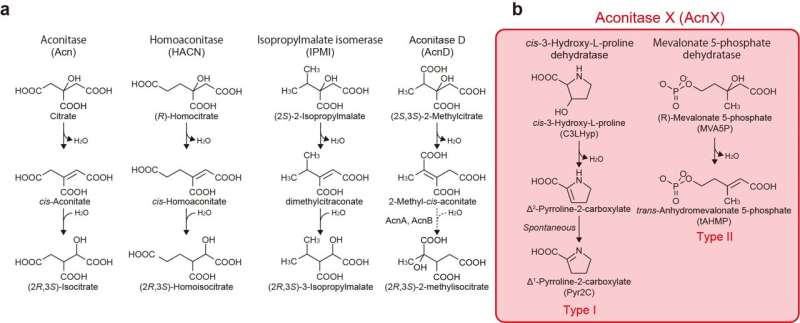
The aconitase superfamily currently contains four functional enzymes including the archetypical aconitase (referred to as "other aconitase enzymes"), and one hypothetical aconitase X (AcnX). The aconitase enzymes catalyze the homologous stereospecific isomerization, and their three-dimensional structures and catalytic mechanisms including the [4Fe-4S] iron-sulfur cluster are very similar each other (Fig. 1a). Therefore, the aconitase superfamily (enzymes) is a typical example that is suitable for the so-called "recruitment hypothesis of enzyme evolution"; the gene duplication of multi-specific enzymes, followed by the narrowing of substrate specificity (ref. 1).
AcnX (subfamily) is further classified into AcnXType-I consisting of a single polypeptide, and AcnXType-II consisting of (fragmented) small and large polypeptide chains. In 2016, we first revealed that AcnXType-I enzyme from bacteria functions as a cis-3-hydroxy-L-proline (C3LHyp) dehydratase (Fig. 1b) (ref. 2). Furthermore, in 2018, other researchers reported that AcnXType-II enzyme from archaea functions as a mevalonate 5-phosphate (MVA5P) dehydratase (ref. 3). To elucidate their catalytic mechanisms, we herein report for the first time the crystal structures of AcnXType-I from Agrobacterium tumefaciens, a plant pathogenic bacterium (AtAcnX), and AcnXType-II from Thermococcus kodakarensis, a hyperthermophilic archaeon (TkAcnX).
AtAcnX and TkAcnX commonly consisted of four domains (fragments), and their structural frameworks of each domain were similar to their counterparts of other aconitase enzymes (Fig. 2). TkAcnX had a cuboidal [3Fe-4S] cluster, which must be derived from the [4Fe-4S] cluster unit via the loss of one iron atom, similar to other aconitase enzymes (Fig. 3a, b). Surprisingly, AtAcnX had a planar [2Fe-2S] cluster (Fig. 3c, d). Most interesting question was whether AtAcnX and TkAcnX can recognize substrates without structural similarity. Collectively, the (superimposed) backbones of C3LHyp and MVA5P were recognized by homologous residues between AtAcnX and TkAcnX (Fig. 4a), whereas their specific structural moieties by different residues (Fig. 4b, c). Since the former residues are completely conserved in other aconitase enzymes, they must be "most ancestral" active sites for aconitase superfamily. Furthermore, the acyclic MVA5P is structurally similar to those of aconitase enzymes, whereas TkAcnX recognized the substrate through homologous manners to AtAcnX, suggesting that substrate specificities (and [4Fe-4S] clusters) for TkAcnX and other aconitase enzymes had acquired each other independently.
![Figure 2. Overall crystal structures of AtAcnX, TkAcnX, and mAcn (aconitase from bovine mitochondria).These molecules commonly contain the indicated [Fe-S] cluster, and consist of four distinct domains 1 (yellow), 2 (green), 3 (blue), and 4 (red) and a linker region (black). The small subunit of TkAcnX corresponds to fragmented domain 4. In comparisons with AcnX, several large insertions in domains 1-4 of mAcn were colored in ocher, light-green, light-blue, and pink, respectively. Credit: Seiya Watanabe, Ehime University The molecular evolution of the aconitase superfamily](https://scx1.b-cdn.net/csz/news/800a/2021/the-molecular-evolutio-1.jpg)
The common ancestor of aconitase superfamily (open circle in Fig. 5), appearing before the previously proposed one (closed circle), had a similar structural framework and a few residues as active site (described above), whereas there was no [Fe-S] cluster. These results provide novel insights into the evolutionary scenario of the aconitase superfamily based on the recruitment hypothesis, and requirement of complicated metabolic pathways in primordial cell.
-
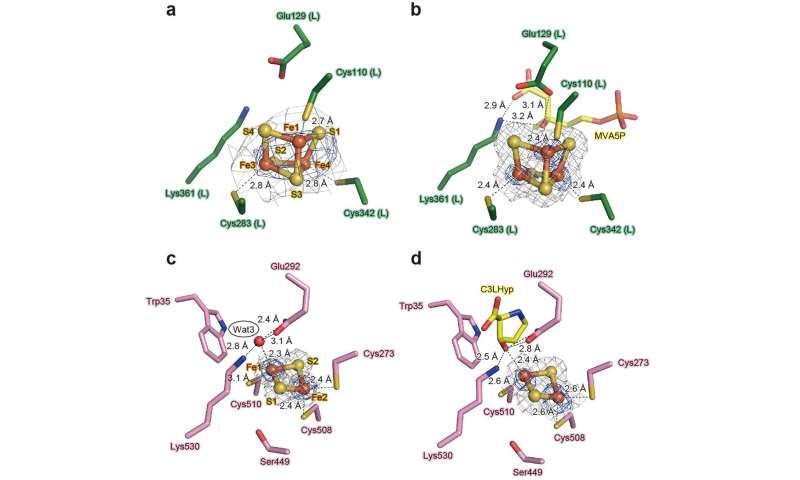
Figure 3. Analysis of the [Fe-S] cluster in AcnX. The observed [3Fe-4S] cluster of TkAcnX in the apo-form (a) and in complex with MVA5P (b). The observed [2Fe-2S] cluster of AtAcnX in the apo-form (c) and in complex with C3LHyp (d). Credit: Seiya Watanabe, Ehime University -
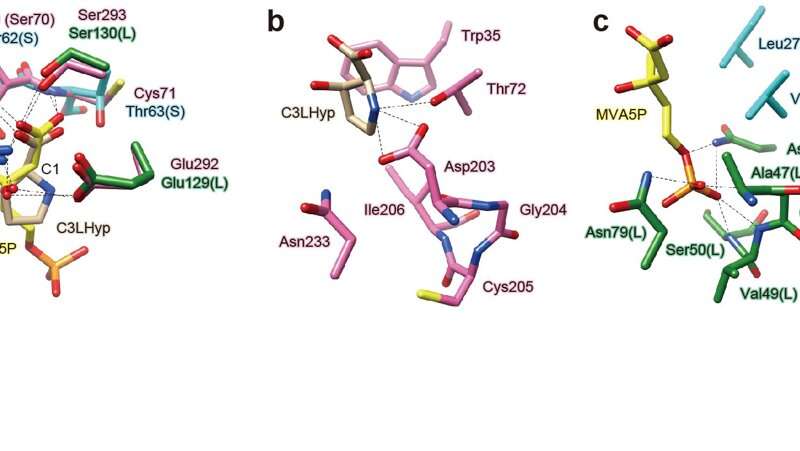
Figure 4. Substrate-binding sites in AcnX.The superposition of active sites that recognize common (a) and different structural backbones (b, c) between C3LHyp and MVA5P. Pick-, light-blue and green residues are derived from AtAcnX, and the small (S) and large subunits (L) from TkAcnX, respectively. Credit: Seiya Watanabe, Ehime University -
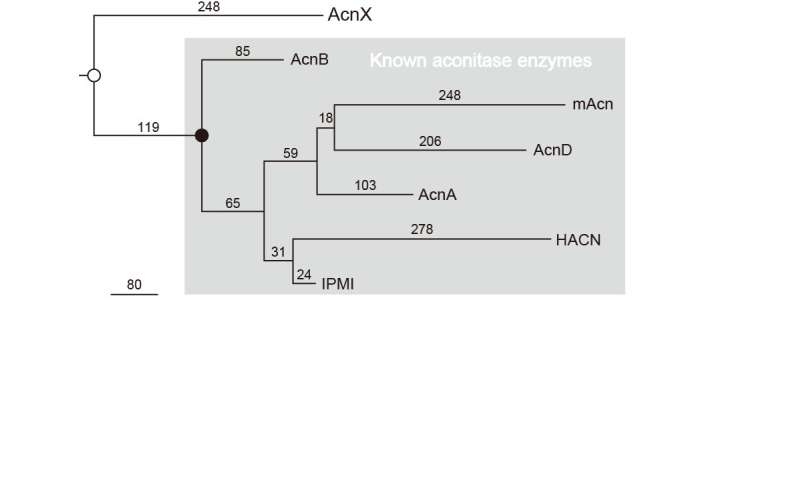
Figure 5. Phylogenetic tree of the aconitase superfamily. The closed circle indicates the common ancestor for other aconitase enzymes that had previously been proposed, which contains the [4Fe-4S] cluster. The open circle indicates the common ancestor for the aconitase superfamily that is proposed in the present study. Credit: Seiya Watanabe, Ehime University
More information: Seiya Watanabe et al, Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily, Communications Biology (2021). DOI: 10.1038/s42003-021-02147-5
Journal information: Communications Biology
Provided by Ehime University