November 13, 2020 feature
Holographic fluorescence imaging to 3-D track extracellular vesicles
Biologists commonly use fluorescence microscopy due to the molecular specificity and super-resolution of the technique. However, the method is withheld by imaging limits. In a new report on Science Advances, Matz Liebel and a research team at the Barcelona Institute of Science and Technology and the Massachusetts General Hospital in Spain and the U.S. reported an imaging approach to recover the full electric field of fluorescent light using single-molecule sensitivity. The team experimented with the concept of digital holography for fast fluorescence detection by tracking the three-dimensional (3-D) trajectory of individual nanoparticles using an in-plane resolution of 15 nanometers. As proof-of-concept biological applications, the researchers imaged the 3-D motion of extracellular vesicles inside live cells.
Nano delivery in living tissue
In this work, Liebel et al. developed fluorescence holography-based 3-D particle localization across extracellular vesicles inside live cells and observed strongly confined vesicles with periods of active transport. Delivering cargo transport in vivo is presently a significant challenge, in order to actively implement minimally invasive nanomedicine platforms. Nanoparticles (NPs) and extracellular vehicles can be engineered as promising candidates to deliver as vehicles but scientists do not yet understand the precise journey of such devices in living tissue.
To overcome these challenges, they must develop wide-field three-dimensional (3-D) single-particle imaging methods to track individual particles simultaneously as they travel to their intended destination. Research teams had previously implemented holographic approaches to microscopy, although the incoherence of fluorescent light is not well suited for live cells or single-molecule imaging. In comparison, shearing interferometry is a promising method to accomplish single shot recording of dynamic processes. The underlying idea behind shearing interferometry includes self-interference to access phase gradients down to a single photon level to achieve single shot fluorescence holography. The mechanisms developed in this work therefore serve to observe intracellular translocation across micrometer length scales to provide biologists with deeper insight to intracellular mechanisms.
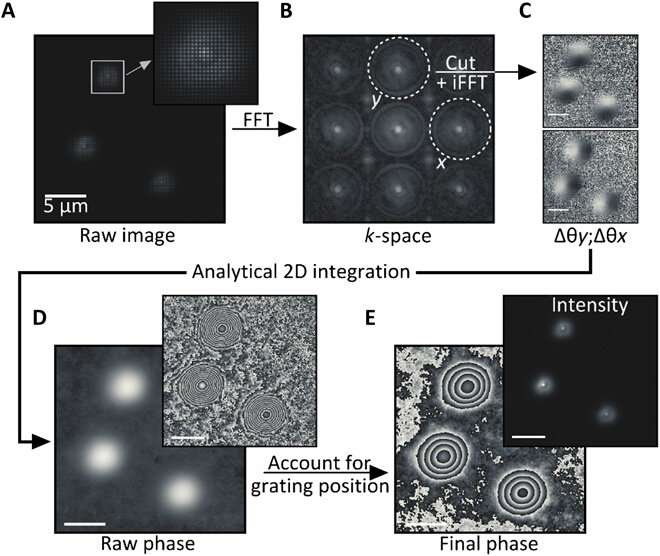
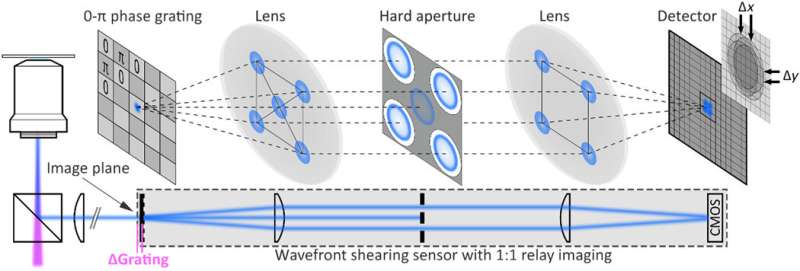
The team used a wide-field fluorescence microscope with a wavefront shearing sensor composed of a relay imaging system. The geometry of the setup ensured that non-zero phase gradients were measured and allowed Liebel et al. to perform single-photon self-interference across an entire image. The team imaged fluorescent polystyrene beads as out-of-focus 200 nm particles and extracted the intensity information as argument modulus of the filtered images for phase gradient extraction. After observing the full electric field, they used Fourier optics to correct complex scattering-induced aberrations or construct images at any plane of choice. The team focused on 3-D localization experiments requiring the recovery of the precise position of an emitter of interest across all dimensions, including the Z-plane. Computational focusing efforts indicated the precise ability to determine the 3-D position of multiple freely diffusing fluorescent particles.
Testing the computational focusing trajectory
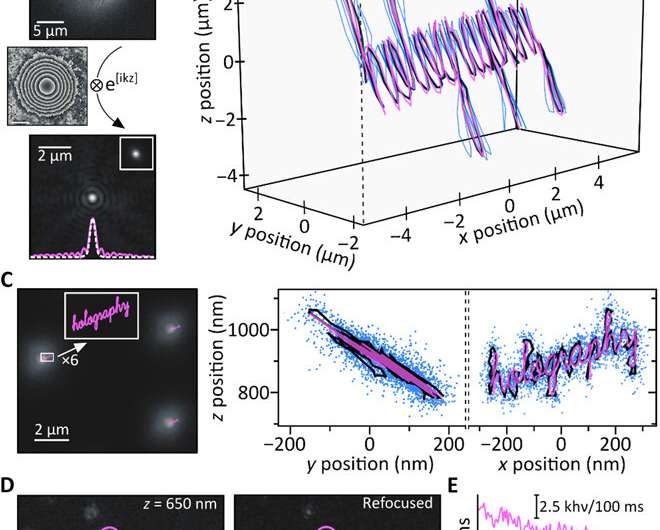
To test computational reasoning behind the setup, Liebel et al. generated a known 3-D trajectory and moved a sample containing immobilized fluorescent beads—while recording images along the path. They recovered the phase and amplitude information and determined the 3-D positions of individual particles using numerical propagation. To quantify the accessible Z-range, they experimentally defocused individual particles and then computationally refocused the images to obtain artifact-free measurements across a Z-range of approximately eight µm. It is important to ensure precise nanoscale localization across micrometer-length scales in 3-D to image diffusing nanoscale particles. Fluorescence holography met these requirements. As proof of concept, the scientists imaged the word "holography," where each individual letter of input measured less than 50 nm in width to obtain a well-resolved output, confirming the super-resolution capacity of fluorescent holography.
Single-molecule imaging and the cellular uptake of nanoparticles
The team showed how fluorescence holography functioned under biologically important super-resolution conditions by measuring a sample composed of individual molecules. Despite markedly reduced fluorescence intensities in the experimental setup, the team obtained computational focusing to the diffraction limit even for photon levels as low as 104 photons. They visualized intracellular trafficking of inorganic nanoparticles and extracellular vesicles using the system. As a model system, they used fluorescently labeled gold nanorods that are inert and therefore without interference with cellular functions to accumulate in the cytoplasm as verified using dark-field images of live cells. The team followed the trajectories of particles by recording time-lapse fluorescence images and extracted the phase and amplitude terms. The widely varying point spread functions (PSFs) indicated the presence of nanorods at different Z-positions relative to the focal plane.
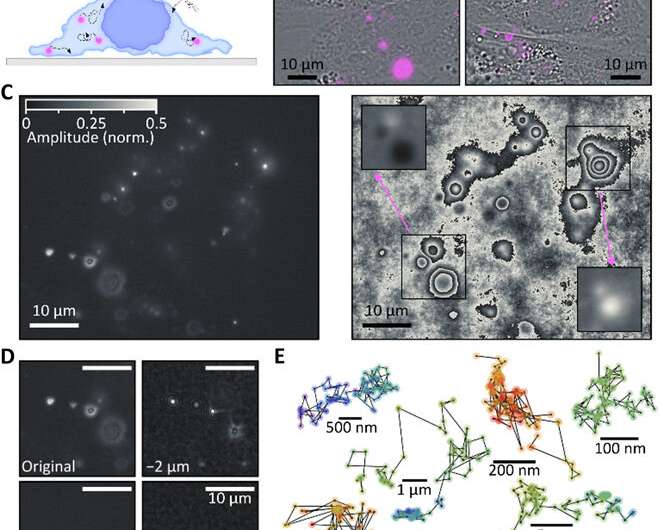
The team performed 3-D localization of each individual nanorod in the cell and reconstructed particle trajectories across 100 frames of observation to obtain six representative categories, where some particles were immobile during the 200 seconds of observation time, while others freely diffused across several micrometers. The remaining particles showed both bound and diffusing states. In this way, the underlying fluorescence holography method could accurately determine 3-D positions.
Cellular uptake and active transport of extracellular vesicles
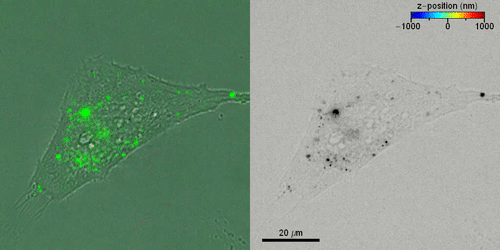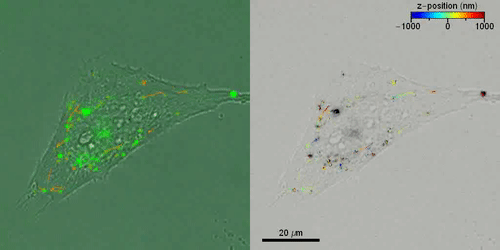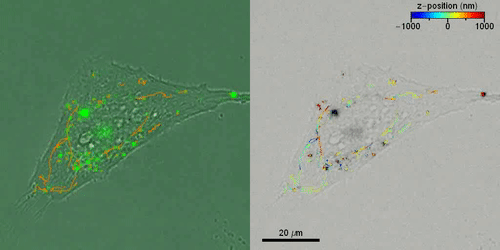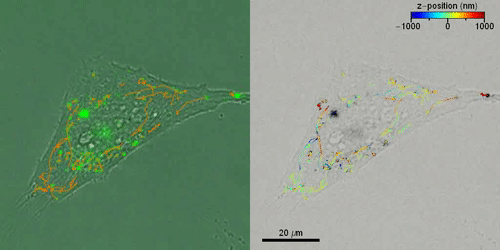
Liebel et al. then studied the active 3-D transport of extracellular vesicles (EVs) inside live cells by incubating HeLa cells with fluorescently labeled EVs. They acquired fluorescent holograms every four seconds to reconstruct 3-D trajectories of individual EVs through a combination of automated and manual trajectories, linking the 3-D EV positions. Liebel et al. overlaid time-lapse amplitude projections of fluorescent holograms with simultaneously recorded bright-field images of individual cells, to show how most EVs were localized at the edge of the adherent cells. The observations and calculations suggested that the EVs were trapped inside an area, confining their motion to a specific volume; most likely belonging to the cellular cytoskeleton.
Outlook
In this way, Matz Liebel and colleagues devised a large field-of-view single-shot fluorescence holography method to allow 3-D single-particle tracking across a Z-range approximating eight micrometers. To prove this concept, the team implemented an easy experimental setup with an optimized photon throughput. The optimized features allowed fluorescence holography to be an ideal approach to study particle tracking in real-time. The team showed 3-D single-particle tracking and observed the motion of nanoscale objects in live cells, such as fluorescently labeled gold nanorods and EVs (extracellular vesicles). While gold nanorods only aggregated in the cytoplasm without internalization in the nucleus, the EVs accumulated at the edges of adherent cells in a crowding effect. Liebel et al. expect to conduct additional staining to identify the intracellular cytoskeleton, thereby connecting the intracellular architecture to the motion of extracellular vesicles. These efforts will shed light on the precise mechanisms of cargo transport and particle internalization inside cells with important applications in nanomedicine to answer critical questions in biology and medicine. The mechanism is equally suited to conduct other volumetric imaging methods to track inside tissues and for biochemical calcium imaging.
More information: Matz Liebel et al. 3D tracking of extracellular vesicles by holographic fluorescence imaging, Science Advances (2020). DOI: 10.1126/sciadv.abc2508
Mercedes Tkach et al. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go, Cell (2016). DOI: 10.1016/j.cell.2016.01.043
D. GABOR. A New Microscopic Principle, Nature (2008). DOI: 10.1038/161777a0
Journal information: Science Advances , Cell , Nature
© 2020 Science X Network