Simulations show new phenomenon with nanopore DNA sequencing
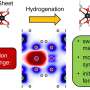
Any truck operator knows that hydraulics do the heavy lifting. Water does the work because it's nearly incompressible at normal scales. But things behave strangely in nanotechnology, the control of materials at the scale of atoms and molecules. Using supercomputers, scientists found a surprising amount of water compression at the nanoscale. These findings could help advance medical diagnostics through creation of nanoscale systems that detect, identify, and sort biomolecules.
The unexpected effect comes from the action of an electric field on water in very narrow pores and in very thin materials. That's according to research by Aleksei Aksimentiev and James Wilson of the Department of Physics at the University of Illinois at Urbana–Champaign. They published their findings in Physical Review Letters, June of 2018.
"We found that an electric field can compress water locally, and that water compression would prevent molecules from being transported through small pores," Aksimentiev said. "This is a very counterintuitive effect, because usually it is assumed that a higher electric field would propel molecules faster through the pore. But because the electric field also compresses water, the outcome would be the opposite. That is, the higher electric field would not allow molecules to pass through." In effect, the water compression generated by the higher electric field pushed DNA molecules away from the nanopore channels.
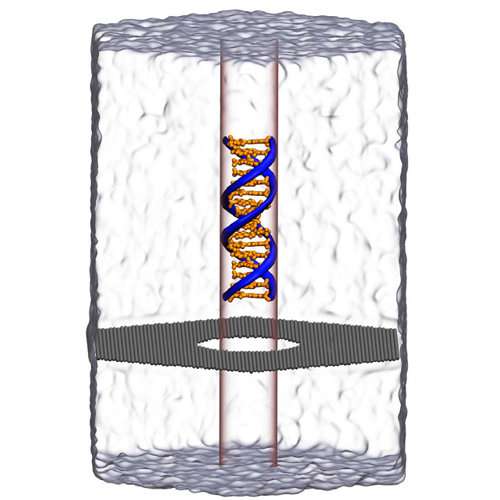
Aksimentiev and Wilson worked with a one-atom-thick graphene membrane. They poked a hole in it 3.5 nanometers wide, just wide enough to let a strand of DNA through. An external electric field pulled the DNA through the hole, like threading a needle. The nucleotide letters A-C-T-G that make the rungs of the double stranded DNA produce signals as they go through the pore, analogous to playing a tape in a tape recorder. This method being developed, called nanopore sequencing, is an alternative to conventional sequencing. It doesn't depend on polymerase chain reaction enzymes to amplify DNA and in theory allows for much longer reads.
"We've been working in the study of nanopore sequencing for some time already, and the goal of the field is to use nanotechnology to read the sequence of DNA, RNA, and proteins directly, without using any kind of enzymes."
Aksimentiev and Wilson were trying initially in the study to quantify how frequently DNA gets captured by graphene pores. Their goal is to increase the capture and in turn the yield of DNA sequenced through the nanopore.
"Surprisingly, we found that as we were increasing this field to increase the rate of DNA capture, we found that it actually doesn't go through after a certain threshold voltage, which was a bit shocking," Aksimentiev said.
"We started looking for all possible things that could go wrong with our simulations," Aksimentiev explained. "We checked everything, and we convinced ourselves that this was indeed a real thing. It's physics speaking to us through all-atom simulations."
They measured the force from the electric field on the DNA molecules, using different DNA constructs and varying the concentration of electrolye solution and the size of the pores and of the membrane. "From these measurements, we came up with this idea that it is water compression that prevents DNA from going through," Aksimentiev said.
Size is everything when it came to the computational challenges of simulating the nanopores. "The problem is that we have to take into account the motion of every atom in our system," Aksimentiev said. "The systems typically are comprised of 100,000 atoms. That was critically important for the discovery of the phenomenon that we have done."
Supercomputer time was awarded through XSEDE, the Extreme Science and Engineering Discover Environment, funded by the National Science. Foundation. XSEDE allocations allowed the researchers use of the Stampede1 and Stampede2 systems at the Texas Advanced Computing Center; and Blue Waters at the National Center for Supercomputing Applications.
Aksimentiev credited XSEDE with a lion's share of the nanoscale study. "I would say that without XSEDE we would not be where we are in our project. Without XSEDE, I don't see how we would be able to accomplish the work that we do. It's not just this project. It's not just this system, but there are so many different systems that our group and other groups are investigating. What I like about XSEDE is that it gives access to diverse systems. The XSEDE portal itself is another benefit, because in one portal I can see everything that happens on all the machines. That makes it very easy to manage allocations and jobs," Aksimentiev said.
"Specifically to Stampede2," Aksimentiev continued, "we were able to run many simulations in parallel. It's not only that our individual simulation uses many cores of Stampede2. At the same time, we also had to run multi copy simulations, where many simulations run at the same time. That allowed us to measure the forces with the precision that allowed us to conclude about the nature of the physical phenomenon. It's been amazing how fast and how accurate the Stampede2 machine works."
James Wilson, a postdoctoral researcher working with Aksimentiev, added that " by running the simulations on Stampede2, I was able to finish twenty simulations in a couple of days, cutting down my time to solution immensely." He explained that just one NAMD molecular dynamics simulation would take about two weeks on local workstations.
"The most important thing," Aksimentiev said, "is that highly accurate, precise simulations on big computers is a discovery tool. This work truly attributes to it, because we set out to do something else. We discovered a new phenomenon in nanopores. And we explain it through simulations. There's so many discoveries to be made with computers. That's why supercomputer research is worth funding."
The next step in this work, furthered Aksimentiev, is to see if the effect also occurs in biological channels and not just with the graphene membrane. They're also exploring the degree of sorting and separation possible for proteins, the cellular machinery of life. "Already in this paper we show that for one protein, we were able to differentiate variants. We'd like to apply it to more complex systems and also find conditions where the effect manifests at lower fields, which would expand its application to detection of biomarkers," Aksimentiev said.
The study, "Water-Compression Gating of Nanopore Transport," was published June of 2018 in Physical Review Letters.
More information: James Wilson et al. Water-Compression Gating of Nanopore Transport, Physical Review Letters (2018). DOI: 10.1103/PhysRevLett.120.268101
The authors are Aleksei Aksimentiev and James Wilson of the University of Illinois at Urbana–Champaign. This work was supported by grants from the National Institutes of Health (Grants No. R01-GM114204 and No. R01 HG007406), and through a cooperative research agreement with the Oxford Nanopore Technologies. The authors gladly acknowledge supercomputer time provided through XSEDE Allocation Grant No. MCA05S028 and the Blue Waters petascale supercomputer system (UIUC).
Journal information: Physical Review Letters
Provided by Texas Advanced Computing Center