Researchers develop new CRISPR/Cas process using Japanese ricefish
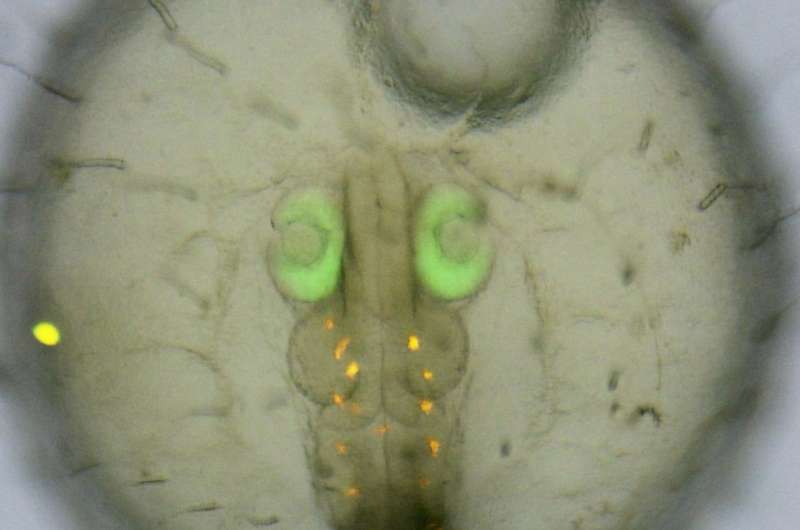
The molecular tool CRISPR/Cas allows introducing DNA double strand breaks into any gene of interest consequently resulting in stochastic mutations at the site of the target gene. However, precise gene repair through the application of a rescue construct suffers from limited efficiency. Researchers at Heidelberg University have now found a solution for this problem. Applying their new approach on the model organism medaka, the researchers laid the groundwork for easily integrating the repair copy of a defective gene into the DNA. As developmental biologist Prof. Dr. Joachim Wittbrodt explains, this efficient process makes precise genome editing possible in basic research, bringing the tool much closer to its application in medical treatment. The research results were published in eLife.
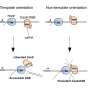
Gene editing involves finding and targeting the precise location in the genome that causes gene mutations. A guide RNA is constructed for this task. It consists of RNA sequences that are complementary to the target DNA sequence. The guide RNA docks at the desired location of the DNA where the CRISPR/Cas as a pair of molecular "scissors" cuts the double strand. The cell's own repair system then jumps into action. Individual DNA building blocks can get lost upon repair of these breaks, causing stochastic mutations in selected target genes. According to Prof. Wittbrodt of Heidelberg University's Centre for Organismal Studies, this method has been used successfully in virtually every organism studied.
However, routine use of precise editing that result in exactly defined modifications in any gene remained elusive so far. As Dr. Arturo Gutierrez explains, the cell's own repair system, which quickly closes the breaks in the double DNA strand, is to blame. Whereas this mechanism, known as "Non-Homologous End Joining" (NHEJ), is not a problem for efficiently introducing stochastic genetic modifications, it competes with a second, extremely precise repair process called "Homology Directed Repair" (HDR). Just like when a replacement part is installed, both ends have to fit perfectly, so that HDR can replace the defective gene with the correct repair copy. "Unfortunately, NHEJ connects the copies introduced into the cells into long continuous chains, rendering them unusable," says Tinatini Tavhelidse.
Using the Japanese ricefish medaka, Prof. Wittbrodt's team developed and validated a new approach that enables highly efficient, precise genetic repair, which is a basic prerequisite for application in gene surgery. The Heidelberg researchers pursued a simple idea. Instead of using pharmacological compounds with serious side effects to mitigate the unwanted effects of NHEJ, they modified the repair copy such that it could not be "attacked" and rendered useless. Both ends of the copy are blocked by biotin – a B vitamin – to prevent non-homologous end joining. "This inexpensive process now enables efficient gene repair by precisely inserting a single repair copy," adds Dr. Thomas Thumberger.
More information: Jose Arturo Gutierrez-Triana et al. Efficient single-copy HDR by 5' modified long dsDNA donors, eLife (2018). DOI: 10.7554/eLife.39468
Journal information: eLife
Provided by Heidelberg University