RNA changes aided sunflower's rapid evolutionary transformation, domestication

A new University of Colorado Boulder-led study sheds light on the genetic mechanisms that allowed sunflowers to undergo a relatively rapid evolutionary transition from wild to domesticated in just over 5,000 years.
Sunflowers, prized for their seeds and oil, have long held agricultural value for humans. Wild, ancestral varieties of the common sunflower Helianthus annuus are widely distributed across North America and grow smaller seeds than their domesticated counterparts, which have been selected over time for prominent single flower heads and larger seeds with high quality oil.
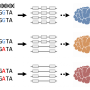
The new study focused on untangling the biological phenomenon of alternative splicing, a regulatory mechanism that allows multicellular organisms to code multiple RNA transcripts and proteins from a single gene. Alternative splicing creates useful efficiencies, but also introduces variation over time. The origins and contributions of alternative splicing to major evolutionary transitions—especially over short periods of time—remain largely unknown.
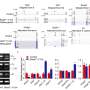
Researchers used RNA sequencing to compare a domesticated H. annuus variety with a wild variety of the same species, focusing on 226 clear cases of splicing differentiation. They were able to identify the genetic regulation of splicing for 134 distinct genes, and found patterns suggesting that this mechanism underlies important domestication traits that have manifested under strong human-induced breeding.
"We were surprised to find that splicing differences were attributable to relatively few regulatory changes" said Chris Smith, a graduate researcher in CU Boulder's Department of Ecology and Evolutionary Biology (EBIO) and the lead author of the study. "We expect that further down the road, various other species could be analyzed this way, too."
The findings, which could have broader implications for agricultural production and scientists' understanding of genetic divergence, were published today in the journal Proceedings of the National Academy of Sciences.
More information: Chris C. R. Smith el al., "Genetics of alternative splicing evolution during sunflower domestication," PNAS (2018). www.pnas.org/cgi/doi/10.1073/pnas.1803361115
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Colorado at Boulder