Deadliest human malaria parasite reveals the genomic chinks in its armor
For the first time, scientists have revealed the essential genes for the most deadly human malaria parasite, Plasmodium falciparum. Researchers from the Wellcome Sanger Institute and the University of South Florida (USF) created new genomic techniques to analyse every gene in the parasite and determine which ones are indispensable.
Published in the journal Science, the core repertoire of genes identified will help researchers to identify and prioritise new drug targets to combat this deadly form of malaria.
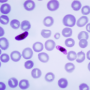
Malaria is caused by Plasmodium parasites and more than 200 million people were infected and nearly half a million people died worldwide from the disease in 2016, predominantly children under the age of five.
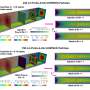
To understand which genes the parasite needs, the researchers disrupted almost every one of the parasite's 5,400 genes. Surprisingly, they discovered that over half of these genes were essential for the parasite to grow in red blood cells.
Researchers at the Sanger Institute carried out a related study last year using the mouse malaria parasite Plasmodium berghei, but the deadly human parasites needed a different approach. The collaborative team used a specialised technique called piggyBac-transposon insertional mutagenesis to inactivate genes at random, and then developed new DNA sequencing technology to identify which P. falciparum genes were affected.
Professor John Adams, one of the study's senior authors and director of USF's Center for Global Health and Infectious Disease Research, said: "What our team has done is develop a way to analyse every gene in this parasite's genome. Using our genetic analysis tools, we're able to determine the relative importance of each gene for parasite survival. This understanding will help guide future drug development efforts targeting those essential genes."
P. falciparum is responsible for half of all malaria cases in the world and causes roughly 90 percent of the fatalities, making it the most lethal malaria parasite in existence. Malaria is a treatable disease when caught early enough, but current antimalarial drugs are failing in many areas due to increasing drug resistance and new drugs are badly needed.
The researchers made more than 38,000 mutations then looked for genes that hadn't been changed, implying they were essential for the parasite to grow. They found more than 2,600 of these essential genes, about 1,000 of which are conserved in all Plasmodium species and have completely unknown functions.
Dr Iraad Bronner, an author on the paper from the Wellcome Sanger Institute, said: "Malaria parasites are extremely technically difficult to manipulate and sequence, and until this study only a few of P. falciparum's essential genes had been determined. Our technological advances enabled us to identify all the essential genes in P. falciparum, the first time this has been possible for a human malaria parasite."
Plasmodium parasites have evolved resistance to many drugs over the last decades, and the spread of parasites resistant to the current front-line antimalarial drug, artemisinin, is an emerging threat. How artemisinin kills parasites is still unknown, but it is thought that resistance could be linked to the proteasome pathway that degrades proteins in the cell. Many of the essential genes found in this study were in the proteasome pathway making it a good target for overcoming artemisinin resistance.
Dr Julian Rayner, one the study's senior authors and Senior Group Leader at the Wellcome Sanger Institute, said: "We need new drug targets against malaria now more than ever, since our current antimalarial drugs are failing. This is the first large scale genetic study in the major human malaria parasite, P. falciparum, and gives a list of 2,680 essential genes that researchers can prioritise as promising possible drug targets. We hope this functional genomics approach will help to speed up the pipeline to develop new treatments for this devastating disease."
More information: M. Zhang el al., "Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis," Science (2018). science.sciencemag.org/cgi/doi … 1126/science.aap7847
J. White el al., "Indispensible malaria genes," Science (2018). science.sciencemag.org/cgi/doi … 1126/science.aat5092
Journal information: Science
Provided by Wellcome Trust Sanger Institute