Computational strategies overcome obstacles in peptide therapeutics development
New computational strategies reported this week in Science might help realize the promise of peptide-based drugs. Peptides are similar to protein molecules, but differ in their smaller size, structure and functions.
Macrocyclic peptides have sparked pharmaceutical industry interest, because they have certain physical and chemical properties that could become the basis of a new generation of medications.
Small peptides have the benefits of small molecule drugs, like aspirin, and large antibody therapies, like rituximab, with fewer drawbacks. They are stable like small molecules and potent and selective like antibodies.
An example of a macrocyclic peptide drug success story is cyclosporine, an immunosuppressant for organ transplants and some autoimmune disorders.
Before the work described in the Science paper, there was no way to systematically design ordered peptide macrocycles like cyclosporine.
Naturally occurring peptides that might serve as reliable starting points, or scaffolds, are few. Equally as frustrating is that they often fail to perform as expected when repurposed. Instead, researchers had resorted to screening large, randomly generated libraries of compounds in the hopes of finding what they needed.
The methods covered in the report, "Comprehensive computational design of ordered peptide macrocycles" now solve these problems.
The lead authors are Parisa Hossienzadeh, Gaurav Bhardwaj and Vikram Mulligan, of the University of Washington School of Medicine Department of Biochemistry and the UW Institute of Protein Design. The senior author is David Baker, professor of biochemistry and head of the institute. Baker is also a Howard Hughes Medical Institute investigator.
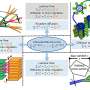
"In our paper," the researchers noted, "we describe computational strategies for designing peptides that adopt diverse shapes with very high accuracy and for providing comprehensive coverage of the structures that can be formed by short peptides."
They pointed out the advantages of this new computational approach:
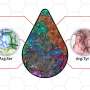
First, they were able to design and compile a library of many new stable peptide scaffolds that can provide the basic platforms for drug candidate architecture. Their methods also can be used to design additional custom peptides with arbitrary shapes on demand.
"We sampled the diverse landscape of shapes that peptides can form, as a guide for designing the next generation of drugs," the researchers said.
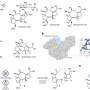
Key to control of the geometry and chemistry of molecules was the design of peptides with natural amino acids, called L-amino acids, and their mirror opposites containing D-amino acids. (The L and D stand for Latin words for rotating to the left or the right, as some molecular structures can have left-or-right handedness or chirality).
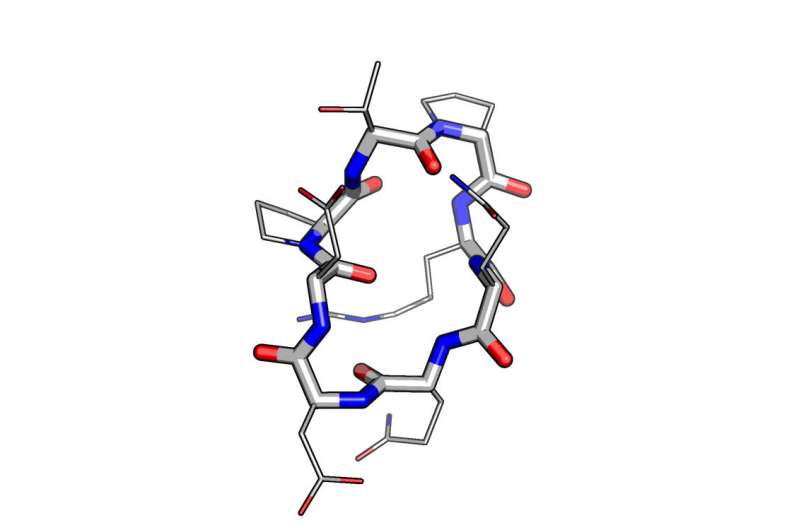
The D-amino acids improved pharmacological properties by increasing resistance to natural enzymes that breakdown peptides. Inclusion of D-amino acids in designs also allows for a more diverse range of shapes.
Designing peptides takes intensive computer power, resulting in expensive calculations. The researchers credited a cadre of citizen scientists and volunteers who donated their spare cellular smartphone minutes and computer time. The Hyak Supercomputer at the University of Washington also ran some of the programs.
The researchers pointed to future directions for their peptide computational design approaches. They hope to design peptides that can permeate cell membranes and go inside living cells.
In other aspects, they plan to add new functionalities to peptide structures by stabilizing the binding motifs at protein-protein interfaces for basic science studies. For clinical applications, they anticipate using their methods and scaffolds for developing peptide-based drugs.
More information: Parisa Hosseinzadeh et al, Comprehensive computational design of ordered peptide macrocycles, Science (2017). DOI: 10.1126/science.aap7577
Journal information: Science
Provided by University of Washington