Chemists brought folded proteins to life

Scientists from ITMO University in Saint Petersburg and Hebrew University in Jerusalem have found a way to recover a protein structure after its chemical denaturation. The method is based on electrostatic interaction between folded, or denatured, proteins and alumina, which unwrap them. The authors highlight the versatility of the method, which works for both specific molecules and multiprotein systems. Theoretically, this can simplify and cheapen the production of drug proteins for Alzheimer's and Parkinson's. The study appeared in Scientific Reports.
Proteins, especially including enzymes as accelerators of chemical reactions, are the basis of pharmaceutical and food industries. Meanwhile, 80 percent of these substances are lost during synthesis. Influenced by unfavorable factors like strong acids, alkalis or heating, proteins denature, losing their native shape and thus any chemical activity. As a result, researchers seek a universal method for recovering protein structure, which could make production more effective and less costly.
Russian chemists in cooperation with foreign colleagues have proposed a solution to this issue with a technological process that returns protein molecules to their original form after denaturation.
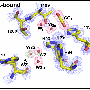
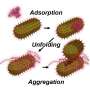
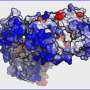
In the new research, the chemists unfolded molecules of three enzymes: carbonic anhydrase, phosphatase and peroxidase. Denatured by a strong alkaline, the proteins were mixed with nanoparticles of alumina in water. Due to electrostatic interaction, the enzymes attracted the nanoparticles and engaged them in forming a supramolecular complex with physical bonds.
This shell of nanoparticles protected the protein molecules from aggregation, enabling the scientists to easily extract them from the aggressive media. Washed from denaturing substances, the enzymes restored their structure by themselves. "Constant exposure of denaturing agents and the tendency of curling macromolecules to aggregation are major obstacles for recovering proteins. When correcting these factors, we were able to regenerate our objects," says Katerina Volodina, second-year graduate student at ITMO University.
Changing the pH, the scientists separated nanoparticles from proteins, showing that the substances involved in the experiment can be repeatedly used.
The authors applied their method to a mixture of two enzymes: carbonic anhydrase and phosphatase (САВ and АсР), renaturating more than half of the molecules, which was an unprecedented result. "Renaturating of multiprotein mixtures is a unique process; it has never been done before. But my colleagues and I believe that further research in this area is in the interest of pharmaceutical companies right now. Theoretically, our method can simplify and cheapen the manufacture of drugs for Alzheimer's or Parkinson's therapy. Many of these medicines are made of proteins," notes Katerina Volodina.
Besides its versatility and high performance, the technology proposed by ITMO University's chemists is also fast and low-cost. The scientists are going to evolve the approach mostly to renaturation of proteins in complex mixtures.
More information: Katerina V. Volodina, David Avnir and Vladimir V. Vinogradov (2017), Alumina nanoparticle-assisted enzyme refolding: A versatile methodology for proteins renaturation, Scientific Reports, www.nature.com/articles/s41598-017-01436-6
Journal information: Scientific Reports
Provided by ITMO University