New insights into ubiquitin signalling
Scientists at the University of Würzburg have generated new insights into the intricate molecular underpinnings of ubiquitin signaling. Their results may provide new avenues for cancer therapy.
The small protein ubiquitin regulates many physiological and pathophysiological processes in the human body. It lives up to its name quite literally by being ubiquitous, both in terms of its abundance and its far-reaching regulatory impact. How ubiquitin exerts its diverse functions is intensely studied all over the world. Finding answers to this question is essential to exploit the ubiquitin system efficiently for therapeutic purposes. Researchers from Würzburg have taken a key step toward this goal. Their results reveal new ways of regulating a ubiquitin ligase.
Enzymes that determine a protein's fate
"Ubiquitin ligases are enzymes that decorate cellular target proteins with ubiquitin and thus determine the fate of these target proteins," says Dr. Sonja Lorenz, senior author on the study. Ubiquitin can act as a "molecular postal code" to guide target proteins to specific locations in the cell, lead them to serve distinct functions, carry molecular signals, integrate into large complexes, or even be destroyed.
Sonja Lorenz heads a research group at the Rudolf Virchow Center for Experimental Biomedicine at the University of Würzburg. Her team and colleagues study a particular ubiquitin ligase, HUWE1, that has been ascribed key roles in tumor formation and is considered a promising, yet unexploited cancer-therapeutic target. Their new results on the molecular mechanism of HUWE1 are reported in the journal eLife.
Divide and rule: breaking down a protein giant
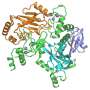
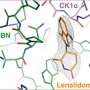
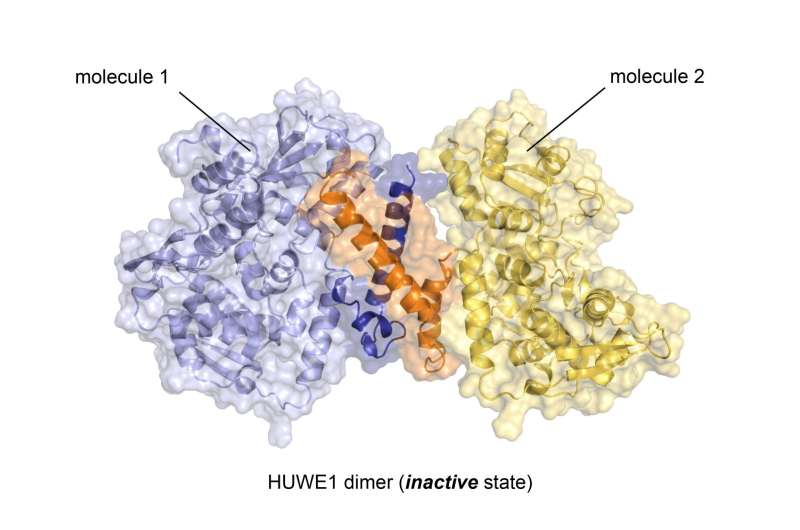
With almost 4,400 amino acids, HUWE1 is an extremely large protein. Its three-dimensional structure, for the most part, is unknown. "The enormous size of HUWE1 and its flexibility present a considerable challenge for structural biologists," says Sonja Lorenz. To get a handle on the protein giant, her research team followed the ancient Roman principle "Divide and rule," and has initially determined the atomic structure of a portion of HUWE1 using X-ray crystallography.
This structure reveals a new and intriguing feature of HUWE1: Two HUWE1 molecules can pair up to form a complex known as a "dimer," thereby shutting down their enzymatic activities.
Imbalances with consequences
How does the cell prevent HUWE1 from forming dimers when the enzyme needs to be active? The Würzburg researchers also provide an answer to this question: HUWE1 exists in a fine-tuned balance of inactive dimers and single, active molecules. "Various cellular factors can regulate this balance," says Sonja Lorenz.
The tumor suppressor protein p14ARF is one such factor. It inhibits HUWE1, but is frequently lost in cancer cells. The new study provides the first mechanistic explanation of how p14ARF inhibits HUWE1. "The effects of p14ARF on the structure and activity of HUWE1 are extremely exciting," says Sonja Lorenz. "They open up a range of possibilities to manipulate HUWE1 activity that we are following up on."
More information: Bodo Sander et al, A conformational switch regulates the ubiquitin ligase HUWE1, eLife (2017). DOI: 10.7554/eLife.21036
Journal information: eLife
Provided by University of Würzburg