Researchers develop the first platform for DNA simulations
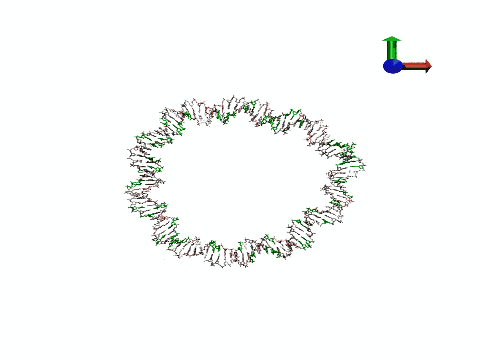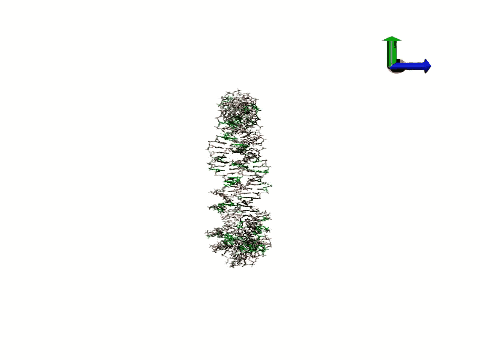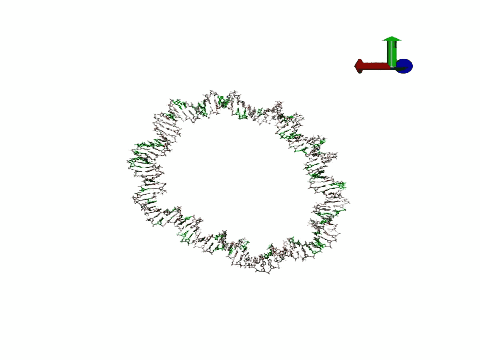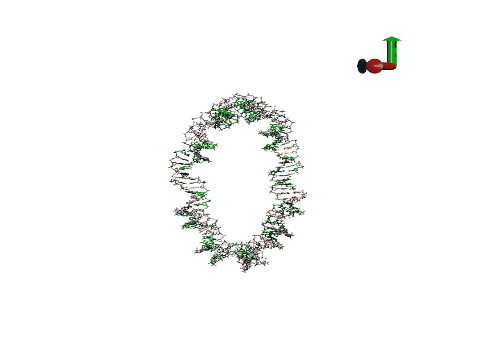
Molecular dynamics is a technique that allows the simulation of DNA movement, including its folding into double, triple or quadruple strands, and even its interaction with proteins and drugs. Molecular dynamics is used to address the processes that occur over time scales ranging from picoseconds to minutes and it can be used for molecular systems of various sizes, from a few nanometres to a meter.
Headed by Modesto Orozco, the Molecular Modelling and Bioinformatics Lab at the Institute for Research in Biomedicine (IRB Barcelona) is developing several theoretical methods to better understand the behaviour of bio-macromolecules, in particular nucleic acids, on a wide spatial-temporal scale and with a focus on biomedical and biotech applications.
Today, the group has published a new model in Nature Methods that allows atomic-level simulations of DNA dynamics with extraordinary accuracy, an achievement that has taken five years of work and the testing of more than 100 DNA systems.
The data is stored in a public website that currently holds more than four terabytes of information. This website is accessible through the Spanish Institute of Bioinformatics (INB) and the ELIXIR-Excellerate network, the largest collection of life sciences data in Europe, to which IRB Barcelona contributes.
The scientists have developed a set of mathematical functions called a force field that describe the movement of the atoms that form DNA. "A force field has never previously allowed the study of such diverse structures in time scales relevant for understanding biological phenomena," says Pablo Dans Puiggròs, IRB Barcelona researcher and first author of the paper.
In addition, the authors present the first online tool devoted to the simulation of nucleic acids, emphasizing that "it allows to predict DNA properties, which can then be compared directly with experiments". "We hope that this new tool and method will open our work to a wider community of users" says Modesto Orozco, director of the project.
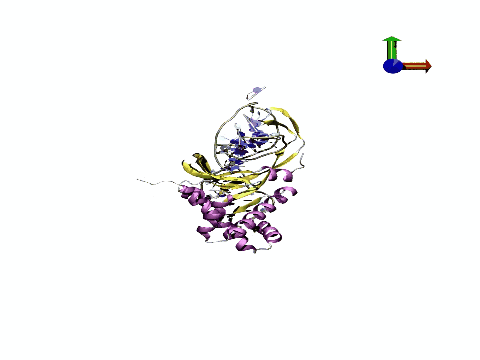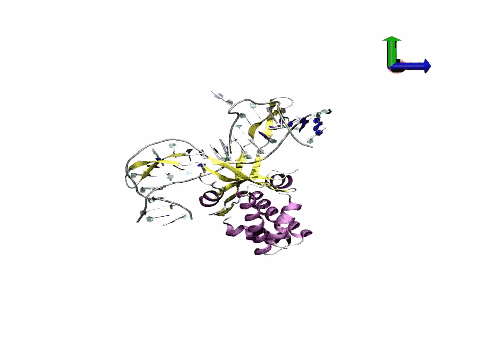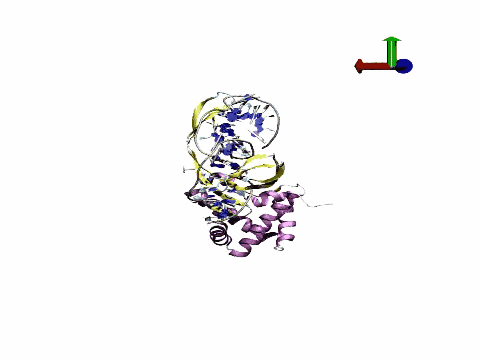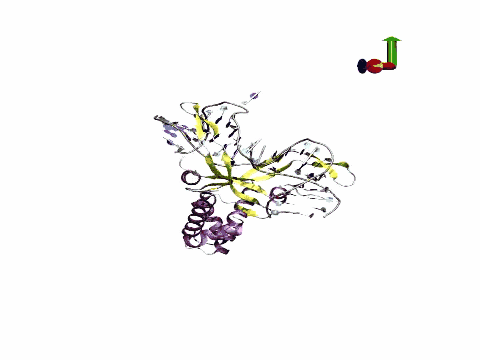
The tool has potential applications in fields ranging from biomedicine to nanotechnology, providing information on mechanisms that underlie DNA regulation and contributing to improvements in the design of drugs that directly or indirectly target DNA. "We are achieving a quantitative leap in the quality of atomic DNA simulations," states Pablo Dans Puiggròs.
"Advances in simulation are bringing us closer to the definition of a theoretical model that will allows us to simulate key aspects of cell life, therefore approaching the dream of describing the behaviour of organisms only based on the basic rules of physics and chemistry," says Modesto Orozco, who is also a senior professor at the University of Barcelona and director of the Life Sciences Department at BSC.
More information: Ivan Ivani et al. Parmbsc1: a refined force field for DNA simulations, Nature Methods (2015). DOI: 10.1038/nmeth.3658
Journal information: Nature Methods
Provided by Institute for Research in Biomedicine